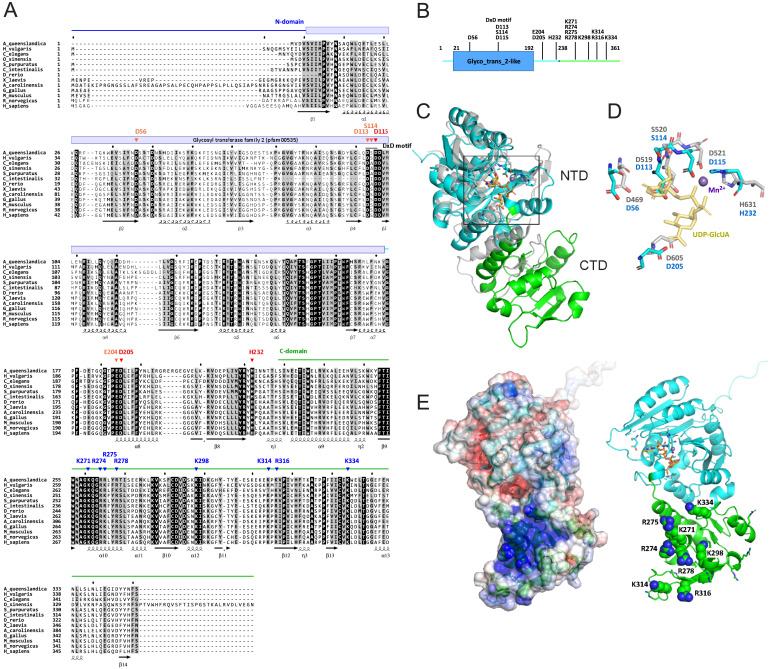
Fig. S10 Sequence alignment and structural characterization of human QTGAL, related to Figure 3 (A) Multiple alignment of 13 QTGAL orthologs by multiple alignment using fast Fourier transform (MAFFT) [S1]. Conserved domains are indicated by colored bars and lines above the alignment. Secondary structures defined from a predicted structure of human QTGAL are indicated below the alignment. Residues predicted to be essential for enzymatic activity are indicated by arrowheads: red, catalytic residues; orange, conserved residues of the catalytic center; blue, basic residues that may interact with the substrate tRNA. (B) Schematic domains of human QTGAL. Color coding is coordinated with (A). Conserved residues involved in catalytic activity or tRNA recognition (shown in A) are indicated. (C) Overall structure of human QTGAL predicted by AlphaFold (https://alphafold.ebi.ac.uk/entry/Q67FW5 ). The N-terminal domain (NTD) and C-terminal domain (CTD) are colored cyan and green, respectively. NTD of QTGAL is superimposed on CTD of K4CP (gray, 2z86 chain D). The catalytic center located in NTD is boxed. (D) Superimposition of the catalytic center of QTGAL and K4CP with a divalent manganese ion and UDP-GlcUA as a sugar donor. (E) (Left) Electrostatic surface potential generated by eF-surf (https://pdbj.org/eF-surf/top.do ) [S2] overlayed on a cartoon model of the QTGAL structure. Positively and negatively charged areas are colored blue and red, respectively. (Right) Conserved positively charged residues (shown in A) are labeled and shown as ball-and-stick models.
Reprinted from Cell, 186(25), Zhao, X., Ma, D., Ishiguro, K., Saito, H., Akichika, S., Matsuzawa, I., Mito, M., Irie, T., Ishibashi, K., Wakabayashi, K., Sakaguchi, Y., Yokoyama, T., Mishima, Y., Shirouzu, M., Iwasaki, S., Suzuki, T., Suzuki, T., Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth, 5517-5535.e24, Copyright (2023) with permission from Elsevier. Full text @ Cell