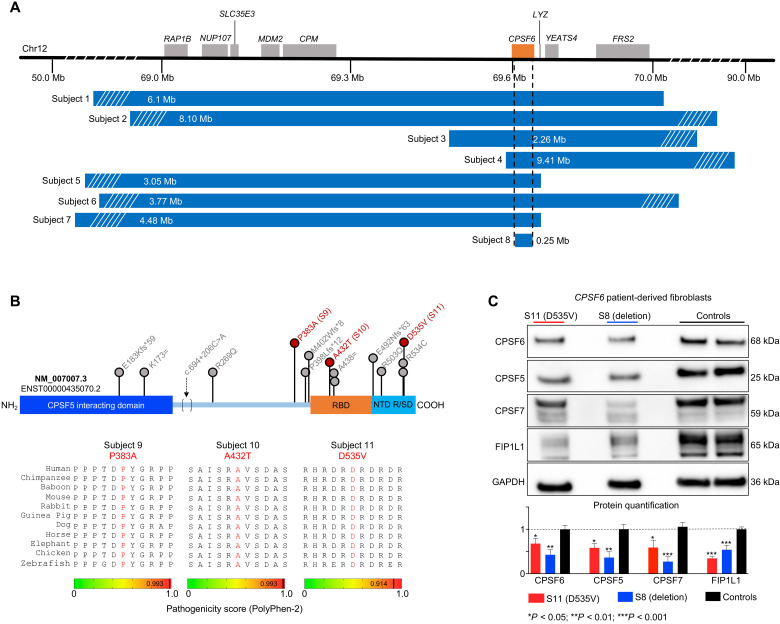
Fig. 1
(A) Deletions spanning CPSF6 (orange box) on chromosome 12q15 identified in eight subjects. Dashed lines indicate the minimal overlapping region, which deleted 99% of CPSF6 (subject 8). Mb, megabases. (B) Schematic of the CPSF6 protein showing the RNA binding domain (RBP; orange), the Arg/Ser-rich domain (R/SD) within the nuclear targeting domain (NTD; turquoise), and the CPSF5-interacting domain (blue). Database searches identified 15 individuals with 13 missense variants (indicated by lollipops), which are plotted here for context, but only subjects 9 to 11 (red) were enrolled in this study. Bottom: Evolutionary alignment shows that the three variants in our subjects affect residues that are conserved from zebrafish to humans and have high pathogenicity scores. See fig. S2 for details of the three splicing variants. (C) Representative Western blot and relative quantification shows that subjects 8 and 11 (the only subjects from whom we obtained fibroblasts) have lower CPSF6, CPSF5, CPSF7, and FIP1L1 protein levels than controls. Data were normalized to GAPDH(glyceraldehyde-3-phosphate dehydrogenase) protein. Data represent means ± SEM from at least four technical and biological replicates compared to healthy age-matched fibroblasts, *P < 0.05, **P < 0.01, and ***P < 0.001.