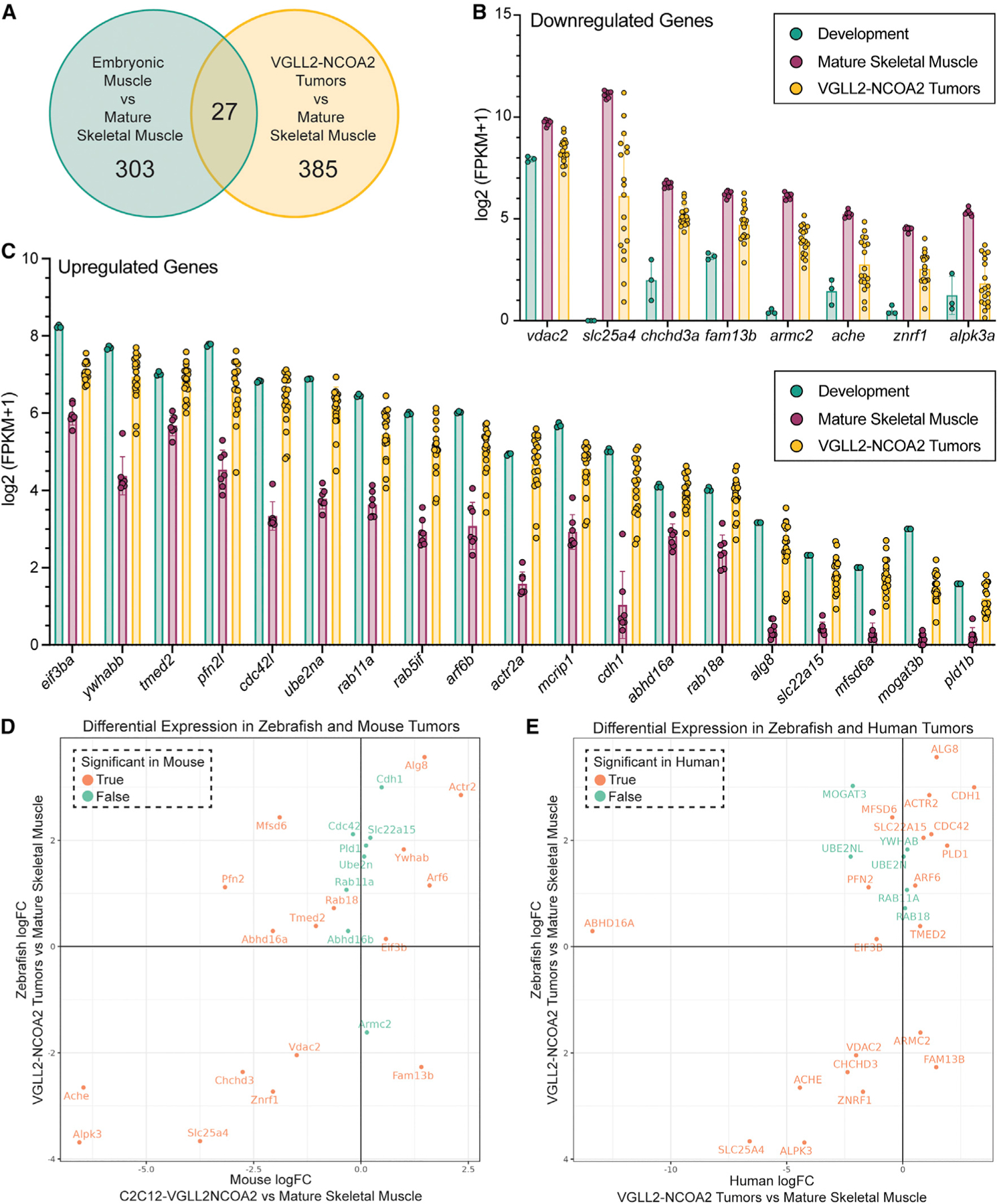
Fig. 4
RNA-seq was performed on n = 18 zebrafish VGLL2-NCOA2 tumors, n = 7 adult zebrafish mature skeletal muscle samples, and n = 3 pooled samples of larval zebrafish from segmentation time points at 1–4 somites, 14–19 somites, and 20–25 somites (10.33, 16, and 19 h post fertilization at 28°C, respectively).
(A) Venn diagram depicting the genes differentially regulated in the developmental time points and tumors compared with mature skeletal muscle, with n = 27 genes differentially regulated in both tumors and development. A false discovery rate (FDR) of 0.01 was used for genes to be included in this analysis.
(B) Plot of FPKM values from n = 8 genes downregulated in developmental time points and tumors compared with mature skeletal muscle. The error bars represent the mean ± SD.
(C) Plot of FPKM values from n = 19 genes upregulated in developmental time points and tumors compared with mature skeletal muscle. The error bars represent the mean ± SD.
(D) Comparison of the expression of these 27 developmental genes in a mouse VGLL2-NCOA2-driven allograft model. Plotted is the fold change of these 27 genes in VGLL2-NCOA2 zebrafish tumors (n = 18) compared with mature skeletal muscle (n = 7), and in C2C12-VGLL2NCOA2 mouse allografts (n = 8) compared with mature skeletal muscle (n = 20; NCBI BioProjects PRJNA625451,28 PRJNA608179,29 PRJNA819493, PRJNA81315330). If the gene is red, it is statistically significant, and, if it is blue, it is not statistically significant in the mouse context.
(E) Comparison of the expression of these 27 developmental genes in human VGLL2-NCOA2 tumors. Plotted is the fold change of these 27 genes in VGLL2-NCOA2 zebrafish tumors (n = 18) compared with mature skeletal muscle (n = 7), and in human VGLL2-NCOA2 tumors (n = 5; Watson et al.4) compared with mature skeletal muscle (n = 803; GTEx version 8). If the gene is red, it is statistically significant, and, if it is blue, it is not statistically significant in the human context. An FDR of 0.1 was used for genes to be included in the analysis in (D) and (E).