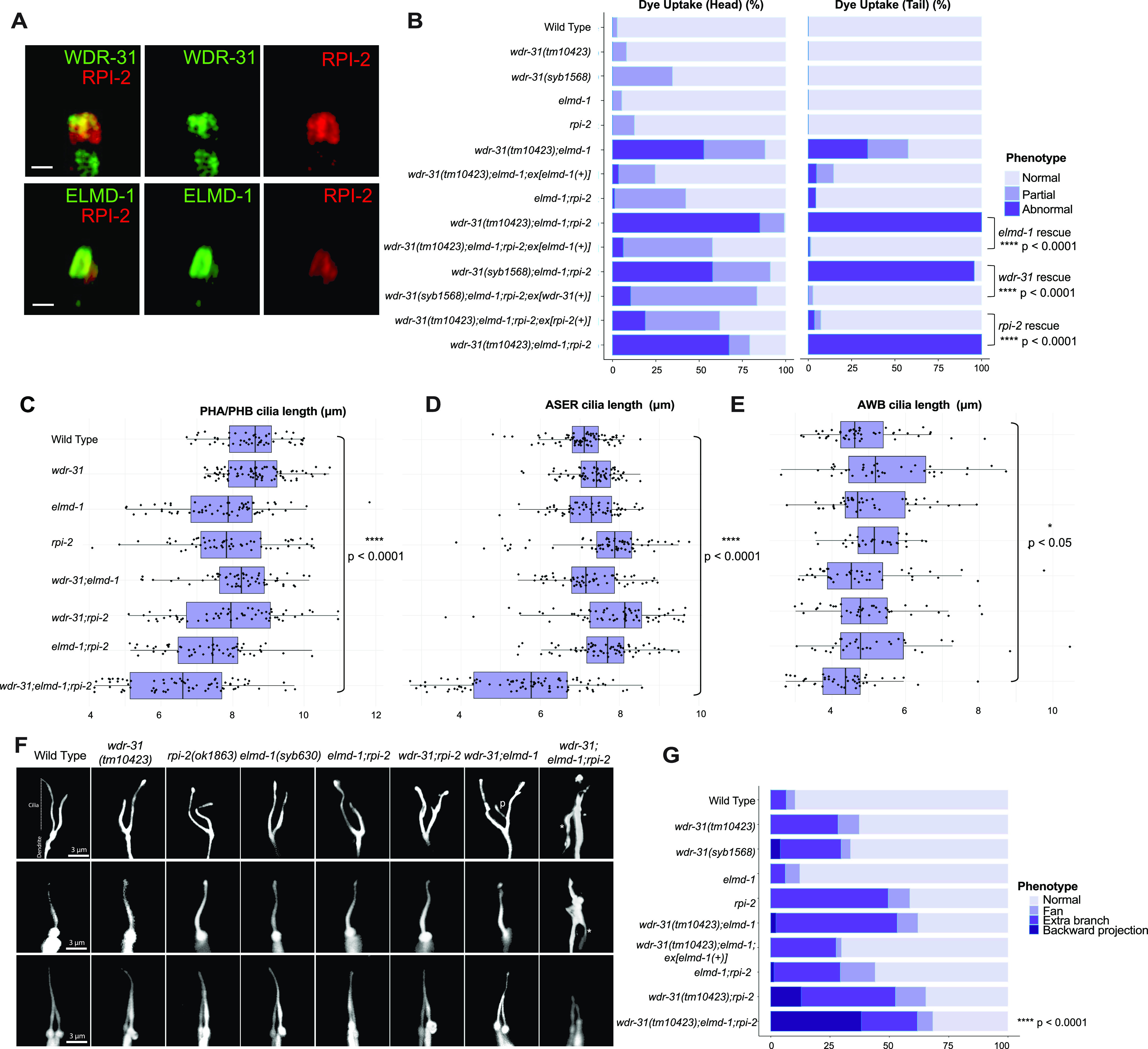
Figure 4.
(A) Shown are co-localizations of RPI-2::mCherry (red, the endogenously labeled RPI-2) with either WDR-31::GFP (green, the endogenously labeled WDR-31) or GFP::ELMD-1 (green, overexpressed) in tail (phasmid) sensory neurons in C. elegans. Scale bars: 1 μm. (B) The fraction of the dye uptake defects is presented in bar charts for WT and the indicated mutants. Fisher’s exact test was performed for statistical analysis between the indicated triple mutants and a rescue gene for Dye assay. Brackets show statistical significance between two strains compared (P < 0.0001 and **** indicate statistical significance). (C, D, E) Shown is the jitter plot for PHA/PHB cilia, ASER cilia, and AWB short cilia length (μm) for WT and indicated mutant strains. Statistical significance between WT and triple mutants was shown with bracket. **** implies statistical significance, which means that P value is lower than P < 0.0001, whereas * means that P-value is lower than P < 0.05. (F) Fluorescence images show the morphology of AWB cilia (fork-like structure located in the head), ASER cilia (amphid channel cilia), and PHA/PHB cilia (phasmid channel cilia) in WT and indicated mutant backgrounds. The backward projection from the cilia is shown with asterisks (*), whereas p indicates the ectopic projections from the middle parts of cilia. Scale bars: 3 μm. (G) The percentage of the abnormality in AWB cilia morphology is depicted in bar charts. Fisher’s exact test was used for statistical analysis of AWB cilia morphology between WT and designated mutants, and **** denotes statistical significance.