Fig. 2
Figure 2. Whole-brain activity maps reveal neural populations encoding self-location
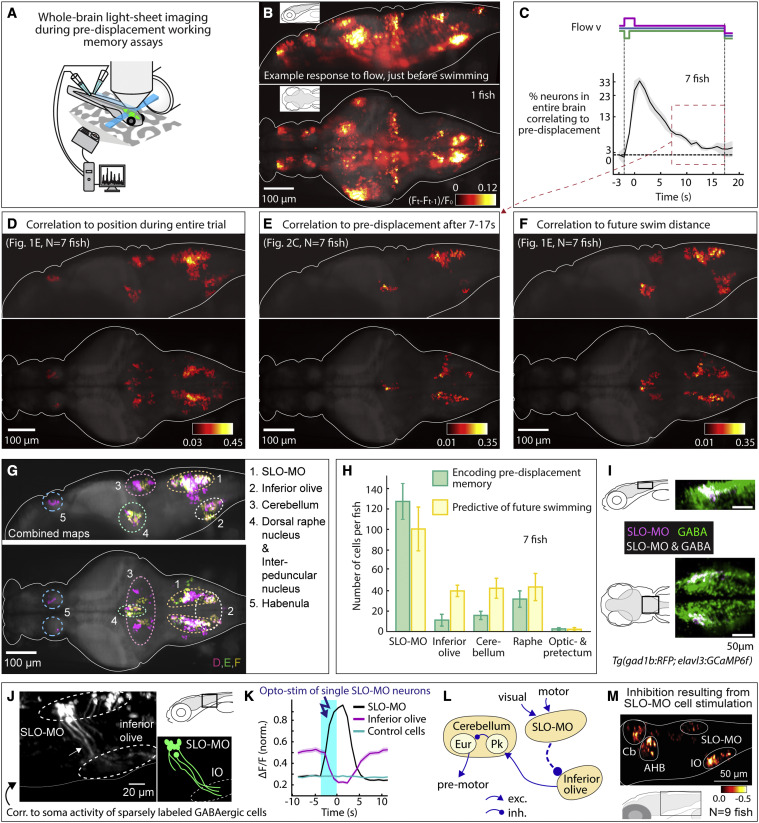
(A) Virtual reality system for paralyzed zebrafish and light-sheet microscope for imaging whole-brain cellular activity.
(B) Example brain-wide activity following forward visual motion, just before swim initiation in Tg(elavl3:GCaMP6f) fish (Figure S5A for earlier and later time points). Shown is the difference between imaging frames F t-1 and Ft normalized by baseline fluorescence, where F t+1 are frames containing the first swim bouts occurring after swim period onset.
(C) Fraction of neurons across the brain with significant correlation to pre-displacement direction as a function of elapsed time. At 17 s after the pre-displacement, a fraction of neurons still encodes pre-displacement direction (shaded regions: SEM; arrow refers to time period used for analysis in (E); backward pre-displacement is shorter than forward to limit swimming during pre-displacement).
(D) Whole-brain map of neurons encoding self-location during pre-displacement, delay, and swim periods. Cells with p < 0.005 for Spearman correlation in at least 80% of time points are shown (STAR Methods). Color bar represents averaged Spearman correlation coefficient to location.
(E) Map of neurons encoding self-location during long delay period (C). Cells with p < 0.005 for Spearman correlation to self-location at every time point between 7 and 17 s are shown. Color bar represents averaged Spearman correlation coefficient of all time points.
(F) Map of neurons whose activity at swim period onset (before first swim bout) predicts total distance swum in swim period. Cells with p < 0.005 for Spearman correlation at first two time points in the swim period are shown. Color bar represents averaged Spearman correlation coefficient.
(G) Combined maps of (D)–(F) listing brain areas containing neurons potentially involved in self-localization.
(H) Numbers of identified neurons (cell segments) per fish per area encoding memory of location during pre-displacement and delay periods, and numbers of neurons predicting future distance swum in the swim period. (Error bars, SEM.)
(I) Dorsal hindbrain map of SLO-MO neurons and GABAergic neurons in Tg(elavl3:GCaMP6f; gad1b:RFP) showing strong overlap between SLO-MO and GABAergic populations (white).
(J) Hybrid functional and anatomical tracing of SLO-MO neurites through sparse expression in Tg(gad1b:Gal4; UAS:GCaMP6f) using fluorescence correlation to SLO-MO cell body activity to help distinguish neurites in sparse gad1b line, showing innervation of IO (STAR Methods).
(K) Optogenetic activation SLO-MO (single neurons of any functional type; STAR Methods) using CoChR59 in Tg(gad1b:Gal4; UAS:CoChR; elavl3:jRGECO1b) (see Figures S7K and S7L) shows IO neurons are inhibited during SLO-MO activation, indicating functional connection consistent with GABAergic inhibition.
(L) Local circuit diagram of hypothesized SLO-MO inhibition of IO (dashed line) and known circuitry from IO to Cb cell types (Pk, Purkinje cells; Eur, eurydendroid cells, homologous to deep cerebellar nuclei).
(M) Hindbrain functional map of decreases in cell activity (color bar, relative decrease ΔF/F) during SLO-MO activation (one neuron at a time) showing activity reduction in IO, cerebellum, anterior hindbrain (AHB), and non-stimulated SLO-MO cells.
Reprinted from Cell, 185, Yang, E., Zwart, M.F., James, B., Rubinov, M., Wei, Z., Narayan, S., Vladimirov, N., Mensh, B.D., Fitzgerald, J.E., Ahrens, M.B., A brainstem integrator for self-location memory and positional homeostasis in zebrafish, 50115027.e205011-5027.e20, Copyright (2022) with permission from Elsevier. Full text @ Cell