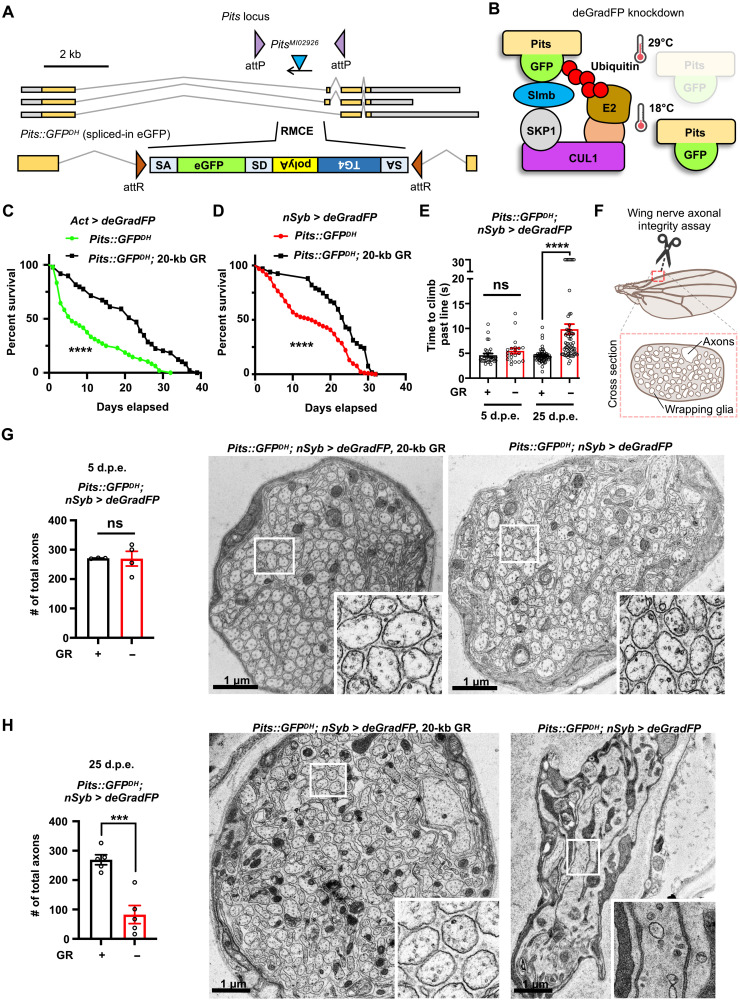
Fig. 2
(A) Schematic of Pits locus in Drosophila with original PitsMI02926 insertion that was converted to Pits::GFPDH by injection of Double Header plasmid (20), resulting in an internally tagged protein trap allele that affects all known Pits transcripts. (B) GFP-tagged fusion proteins can be targeted by deGradFP (UAS-NSlmb::vhhGFP4) to degrade Pits::GFPDH with spatial (GAL4) and temporal (temperature) control. (C and D) Life span is decreased when Pits::GFPDH is ubiquitously (C) or neuronally (D) degraded by deGradFP during development and the adult stage. n is a minimum of 48 flies per genotype (****P < 0.0001). Statistical analyses were determined by the log-rank test. (E) Pits::GFPDH; nSyb-GAL4, UAS-deGradFP flies exhibit progressive climbing defects. Time (seconds) required for flies of the indicated genotypes to climb past 7 cm (n > 20 per genotype). Statistical analyses are one-way ANOVA followed by a Tukey post hoc test. Results are means ± SEM (****P < 0.0001). (F and H) Diagram of the peripheral wing nerves containing peripheral neurons of the anterior wing margin observed with (G and H) transmission electron microscopy. Quantification shows the total axon number at (G) 5 d.p.e. in Pits::GFPDH; nSyb-GAL4, UAS-deGradFP flies containing GR construct (n = 3) and flies without GR (n = 4) and (H) 25 d.p.e. (n = 5 each genotype). Results are means ± SEM (***P < 0.001). Statistical analyses were determined by two-tailed Student’s t test. Scale bars, 1 μm.