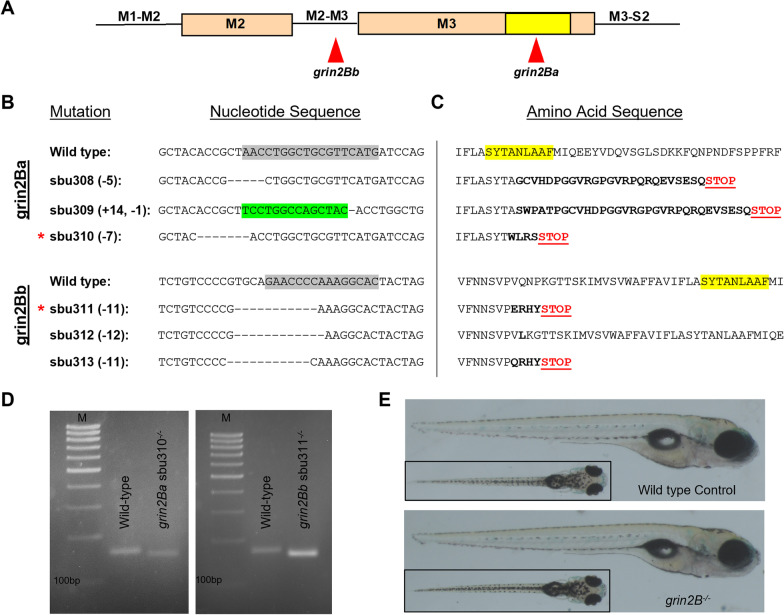
Fig. 2
Generation of loss-of-function lesions in grin2Ba and grin2Bb using CRISPR-Cas9. A Linear representation of the M2 and M3 segments indicating gRNA target sites for grin2Ba and grin2Bb. The SYTANLAAF motif (indicated in yellow) is the most highly conserved motif in iGluRs and is fundamental to their function. B and C Alignments of B nucleotide and C amino acid sequences. gRNA target sites on the nucleotide sequence are indicated (gray highlights). Induced mutations within the nucleotide sequences are denoted as either dashes (deletions) or highlighted green (insertions). Altered amino acid sequences (bolded) and early stop codons (Red STOP) are generated in all alleles except sbu312. Such early translation termination events would prevent encoding the SYTANLAAF motif (yellow highlight) as well as the D2 lobe of the LBD, which would make the receptor non-functional. D Only mutant mRNA is detected for each grin2B lesion by rt-PCR. cDNA amplification of: grin2Ba+/+ and grin2Ba−/− (unless otherwise noted, grin2Ba−/− denotes the sbu310 allele) producing expected product sizes of 158 bp and 151 bp, respectively; and grin2Ba+/+ and grin2Bb−/− (unless otherwise noted, grin2Bb−/− denotes the sbu311 allele) producing expected product sizes of 171 bp and 160 bp, respectively. M denotes marker. For all genotypes, RNA was collected from homozygous intercrosses at 5 dpf. E Lateral and dorsal (inlet) images of representative 6 dpf wild type (top) and grin2B−/− (bottom)