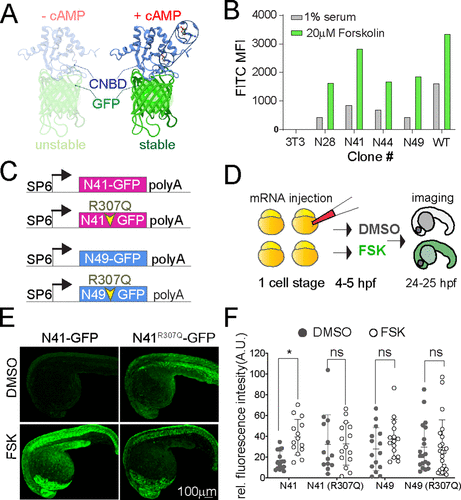
Fig. 1 Figure 1. (A) Ribbon diagram of the cAMP sensor composed of CNBD-GFP protein that is unstable without cAMP but stabilized upon cAMP binding: solution structure of a bacterial cyclic nucleotide-activated K+ channel binding domain in cAMP-free form on the left (PBD 2KXL) and bound to cAMP on the right (PBD 2K0G). (B) NIH 3T3 cells stably expressing CNBD-GFP derived from error-prone PCR were treated with 1% serum or 20 μM forskolin for 17 h. (C) Synthetic mRNA encoding the sensor variants N41 and N49 CNBD and the corresponding cAMP-insensitive controls (R307Q). (D) mRNA was injected into zebrafish embryos at the one-cell stage. Embryos were treated at 4–5 hpf with FSK or DMSO for 20 h, then imaged at 24–25 hpf. (E) Representative images showing 24 hpf embryos injected with N41-GFP and N41R307Q-GFP mRNA at one-cell stage and treated with DMSO or 20 μM FSK starting at 4 hpf. (F) Each dot represents mean GFP intensity of one embryo. Error bars indicate SD: *p < 0.5 by one-way ANOVA (with Šídák’s multiple comparisons), n = 13–22 embryos each condition; ns = not significant. AU = arbitrary unit.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ ACS Chem. Biol.