Image
Figure Caption
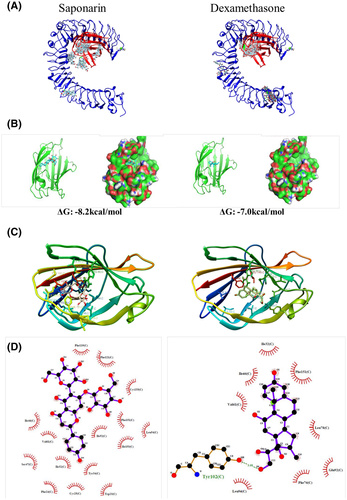
Fig. 10
Molecular docking studies of the interaction between saponarin and TLR4/MD2 complex. (A) Depiction of ligand clusters of saponarin or dexamethasone binding to TLR4/MD2 complex as deduced by protein-ligand docking simulation in Swissdock. (B) Estimated binding energy and pose of the best MD2-ligand binding simulation model. UCSF chimera depicted MD2 protein as a ribbon and provided a surface image. (C) Sidechains of MD2 protein proximal to ligands (within 5 Å) were labeled and visualized by UCSF chimera. (D) 2D image of non-ligand residues and its corresponding atoms of ligand visualized by Ligplot plus.
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ FASEB J.