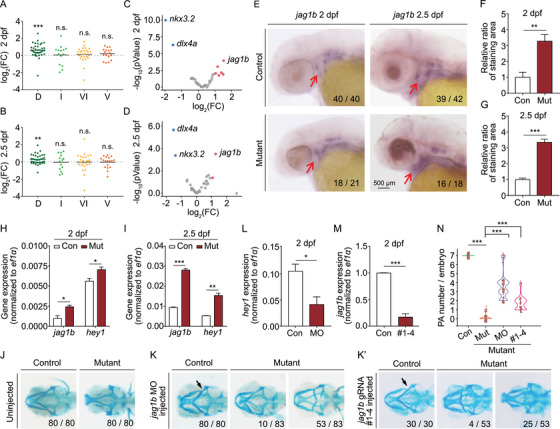
FIGURE 6
The upregulated Jagged‐Notch signaling inhibits PA development in mutants. (A and B) The domain‐specific difference in gene expression in mutants relative to controls measured along the dorsal/ventral (DV) axis at 2 dpf (A) and 2.5 dpf (B). D, dorsal; I, intermediate; VI, ventral‐intermediate; V, ventral. **p < 0.01 and ***p < 0.001. (C and D) Volcano plots summarizing the difference in expression of marker genes in mutants relative to controls measured along with the anterior/posterior (AP) and DV axes at 2 dpf (C) and 2.5 dpf (D). Blue and red dots indicate significantly (p < 0.05) downregulated and upregulated genes, respectively. Gray dots indicate genes that were neither upregulated nor downregulated (p > 0.05). The dlx4a, jag1b, and nkx3.2 genes are indicated. (E–G) Whole‐mount in situ hybridization of jag1b in control and mutant embryos at 2 and 2.5 dpf (E), and summary of the staining area at 2 dpf (F) and 2.5 dpf (G), normalized to the respective controls. (H and I) Summary of jag1b and hey1 mRNA measured in sox10 + cells isolated from control and mutant embryos at 2 dpf (H) and 2.5 dpf (I). (J and K) Representative images of head cartilage staining in control and mutant embryos (J) and embryos injected with the jag1b MO (K) or a set of four CRISPR/Cas9 ribonucleoprotein complexes (K’). Examples of two distinct mutant phenotypes, with the corresponding frequencies, are shown in K and K’. The arrows in K and K’ indicate the characteristic “kink” in the control embryos. (L) Summary of hey1 mRNA measured in sox10 + cells isolated from control embryos and control embryos injected with the jag1b MO. (M) Summary of jag1b mRNA measured in control embryos and control embryos injected with the jag1b CRISPR/Cas9 ribonucleoprotein complexes. (N) Violin plot summarizing the number of PA pairs/embryos in control embryos, mutant embryos, and mutant embryos injected with either the jag1b MO or the jag1b CRISPR/Cas9 ribonucleoprotein complexes