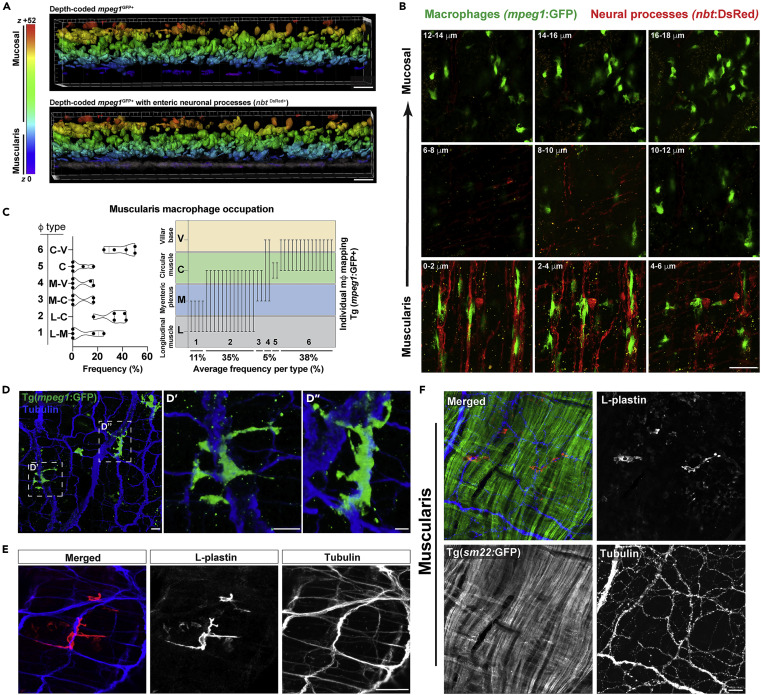
Fig. 3 (A) Volumetric depth-coded analysis (Imaris) of 40 x confocal micrographs from Tg(mpeg1:GFP;nbt:dsRed) ex vivo proximal adult gut highlighting a spatially distinct mpeg1GFP+ MMφ population (purple, top panel) which co-segregate with dense nbtDsReD+ neural processes (gray, bottom panel). Data are representative of at least n = 5 animals. (B) 2 μm confocal z series micrographs (0.5 μm voxel depth with Nyquist sampling) showing intimate mpeg1GFP+ MMφ - nbtDsRed+neural process associations. (C–E) (C, left) Graphical representations of MMφ occupation frequencies spanning the longitudinal muscle – myenteric plexus (L-M), longitudinal muscle – circular muscle (L-C), myenteric plexus – circular muscle (M-C), myenteric plexus – villar ridge base (M-V), circular muscle (C), or circular muscle – villar ridge base (C-V) regions. (C, right) Graphical representations of end-to-end macrophage occupation across gut layers highlighting bridging MMφ phenotype. High-resolution confocal imaging with Nyquist sampling of ex vivo (D) Tg(mpeg1:GFP) adult gut tissue (α- GFP, α-acetylated tubulin immunolabeled) highlight neural bridging of mpeg1GFP+ MMφ processes and (E) wild-type adult gut tissue without Tg (α- L-plastin and α-acetelated tubulin immunolabeled) demonstrating Tg-independent visualization of MMφ-neural processes and interactions. (F) Ex vivo Tg(sm22:GFP) adult gut tissue was fixed and sm22:GFP, L-plastin, and acetylated tubulin were visualized by antibody-mediated immunolabeling to highlight specific MMφ associations with neural processes rather than smooth muscle fibers of the muscularis externa. Scale bars shown = 20 μm (A, F); 50 μm (B, E); 40 μm (D).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ iScience