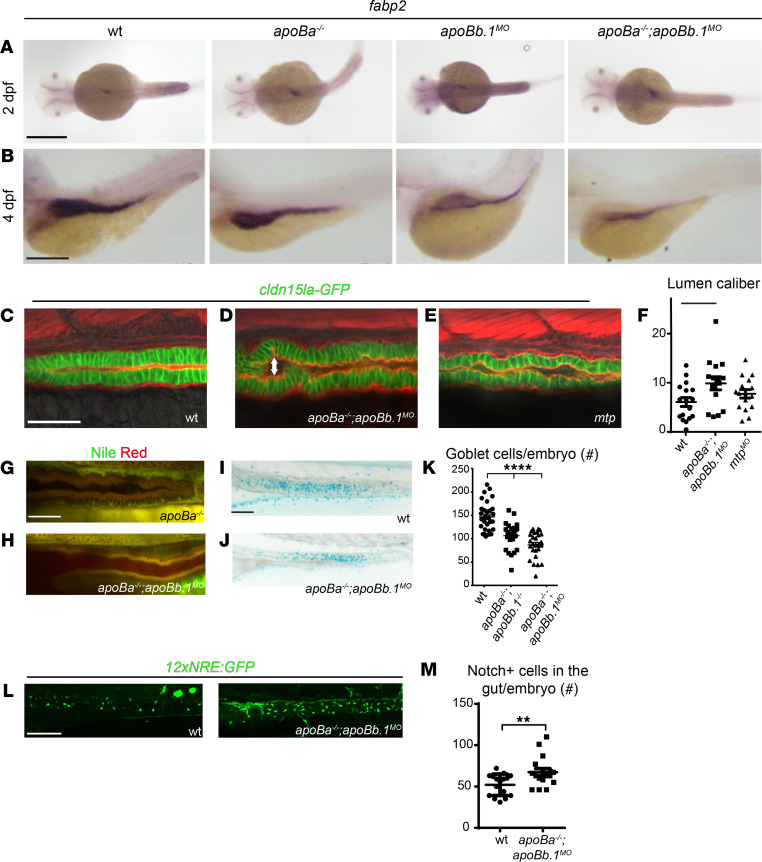
Figure 3 (A and B) WISH showing the spatial expression of fabp2 at 2 (A) and 4 (B) dpf. (C–E) Confocal images at 4 dpf of WT (C), apoBa–/– apoBb.1MO (D), and mtpMO (E) embryos in the TgBAC(cldn15la-GFP)pd1034 reporter background, stained with phalloidin (red). Enlarged lumen caliber (arrow) is detected in apoBa–/– apoBb.1MO and mtpMO embryos, as quantified (F) (n = 2, nWT = 17, napoBa–/– apoBb.1MO = 15, nmtpMO = 16). The data are shown as the mean ± SEM, calculated using ANOVA followed by Tukey’s multiple-comparison test. *P < 0.05. (G and H) Confocal images of the guts of 4 dpf apoBa–/– and apoBa–/– apoBb.1MO embryos stained with Nile red, show no lipid accumulation. (I and J) Bright-field images of 5 dpf embryos stained with Alcian blue showing reduced number of goblet cells in apoBa–/– apoBb.1–/– double mutants (J) as compared with WT embryos (I), as quantified (K) (n = 3, nWT = 30, napoBa–/– apoBb.1–/– = 27, napoBa–/– apoBb.1MO = 27). The data are shown as the mean ± SEM, calculated using ANOVA followed by Tukey’s multiple-comparison test. ****P < 0.0001. (L) Confocal images at 5 dpf of WT and apoBa–/– apoBb.1MO embryos in the Tg(12xNRE:Egfp) reporter background. (M) Quantification of number of cells displaying active Notch signaling in the guts of WT and apoBa–/– apoBb.1MO embryos. The data are shown as the mean ± SEM, calculated using 2-tailed Student’s t test. Scale bar: (A–E, G, and H) 50 μm, (I, J, and L) 100 μm. **P < 0.01. P < 0.05 (considered significant versus control group).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ JCI Insight