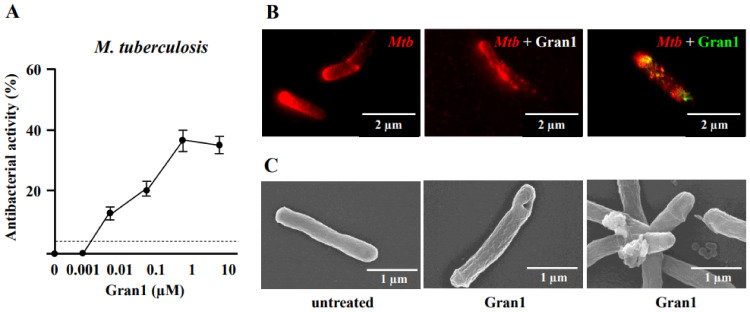
Figure 3
Colocalization of Gran1 with Mtb and cell wall distortion: (A) Extracellular Mtb (2 × 106) were incubated for 72 h with increasing concentrations of Gran1. Afterwards 3H-Uracil was added for 18 h, and uptake was measured by scintillation counting. Data points show the mean antibacterial activity (%) ± SEM calculated from triplicates of twenty independent experiments. Dotted horizontal lines indicate background activity of diluent control. (B) Extracellular Mtb were labelled with NHS-Atto647N and left un-treated (left panel) or were incubated with Gran1 for 30 min (middle and right panel). For signal overlay of mycobacteria and the peptide, Gran1 was labelled using an anti-Gran1 antibody and an Atto594-conjugated secondary antibody (right panel). Samples were acquired by STED microscopy. The middle panel shows Gran-1 treated, NHS-Atto647N-labelled Mtb, and the right panel shows an overlay of NHS-Atto647N and Atto594-labelled Gran1. Representative images of minimum ten examined Mtb per experiment are shown (n = 2). (C) Extracellular Mtb were treated with Gran1 or left untreated for 3 d and processed for scanning electron microscopy. Representative images show untreated Mtb and Gran1-treated Mtb. Images were acquired using a Hitachi S-5200 scanning electron microscope. Magnification 40.000× to 50.000×.