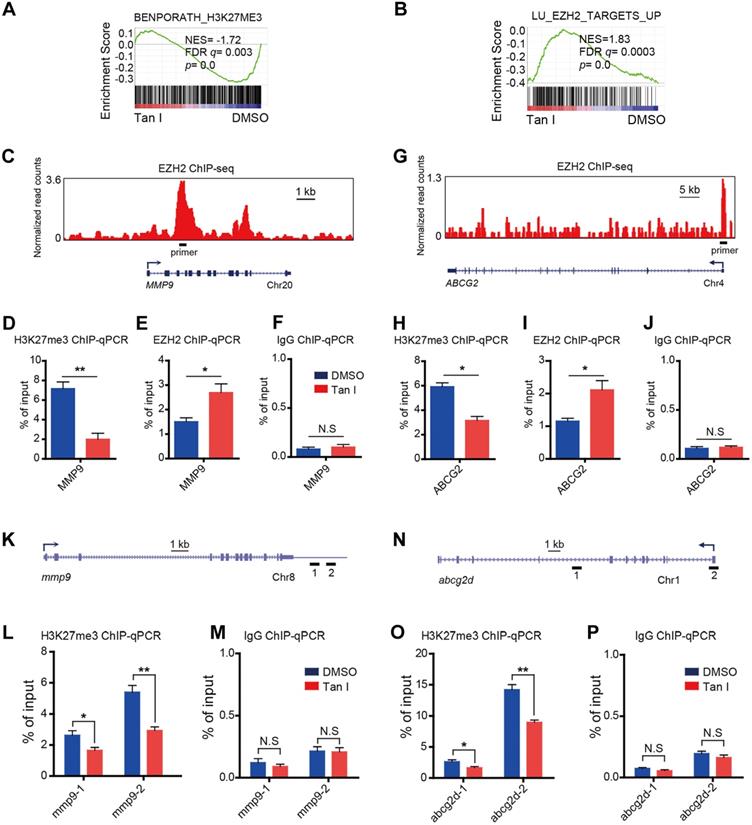
Fig. 6 GSEA of the expression profile of NB4 cells treated with DMSO or Tan I using a histone H3K27me3-associated signature “BENPORATH_H3K27ME3” (A) and an EZH2 target genes-associated signature “LU_EZH2_TARGETS_UP (M2139)” (B). (C) Genome browser track representing the binding sites of EZH2 at MMP9 gene locus in human LNCaP cells. (D-F) ChIP-qPCR assay of H3K27me3 (D), EZH2 (E) or IgG (F) occupancy at MMP9 gene locus in NB4 cells treated with DMSO or 10 µM Tan I for 3 days. (G) Genome browser track representing the binding sites of EZH2 at ABCG2 gene locus in human LNCaP cells. (H-J) ChIP-qPCR assay of H3K27me3 (H), EZH2 (I) or IgG (J) occupancy at the ABCG2 gene locus in NB4 cells treated with DMSO or 10 µM Tan I for 3 days. (K) Horizontal lines with Arabic numeral 1 to 2 indicate the regions at the mmp9 gene locus amplified by qPCR in zebrafish ChIP assay. (L-M) ChIP-qPCR assay for H3K27me3 (L) or IgG (M) occupancy at the mmp9 gene locus in zebrafish embryos treated with DMSO or 60 µM Tan I for 3 days. (N) Horizontal lines with Arabic numeral 1 to 2 indicate the regions at the abcg2d gene locus amplified by qPCR in zebrafish ChIP assay. (O-P) ChIP-qPCR assay for H3K27me3 (O) or IgG (P) occupancy at the abcg2d gene locus in zebrafish embryos treated with DMSO or 60 µM Tan I for 3 days. Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01. All results are from three independent experiments.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Theranostics