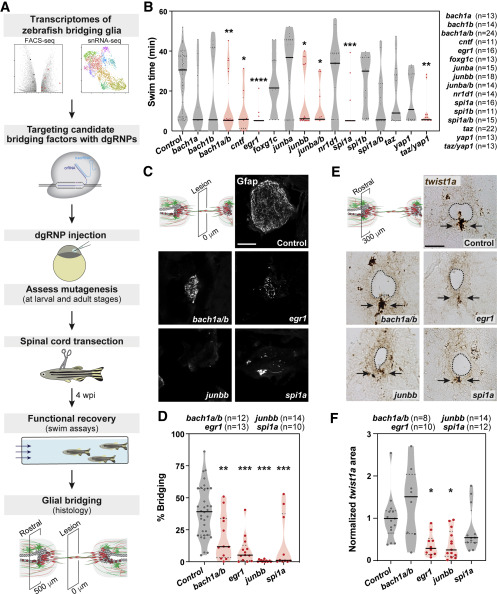
Fig. 4 An EMT-driving gene regulatory network directs glial bridging (A) Pipeline for an in vivo CRISPR-Cas9 screen for glial bridging transcription factors. (B) Functional recovery in CRISPR-Cas9-targeted animals 4 wpi. For each group of targeted animals, uninjected siblings were subjected to SCI and swim assays. Dots represent individual animals. Groups with significantly diminished swim function are shown in red. (C and D) Glial bridging in CRISPR-Cas9-targeted animals. Gfap immunohistochemistry was performed at 4 wpi (C). Representative micrographs show Gfap+ bridges at the lesion site in bach1a;bach1b, egr1, junbb, and spi1a-targeted animals. Percent bridging was quantified for 10–14 animals per group (D). (E and F) twist1a expression in CRISPR-Cas9-targeted animals. RNAscope was performed on SC tissues at 4 wpi (E). Dotted lines delineate central canal edges. Arrows point to ventral ependymal progenitors in distal SC sections. Normalized twist1a was quantified from 8–14 sections per group (F). ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05. Scale bars, 50 μm.
Reprinted from Developmental Cell, 56(5), Klatt Shaw, D., Saraswathy, V.M., Zhou, L., McAdow, A.R., Burris, B., Butka, E., Morris, S.A., Dietmann, S., Mokalled, M.H., Localized EMT reprograms glial progenitors to promote spinal cord repair, 613-626.e7, Copyright (2021) with permission from Elsevier. Full text @ Dev. Cell