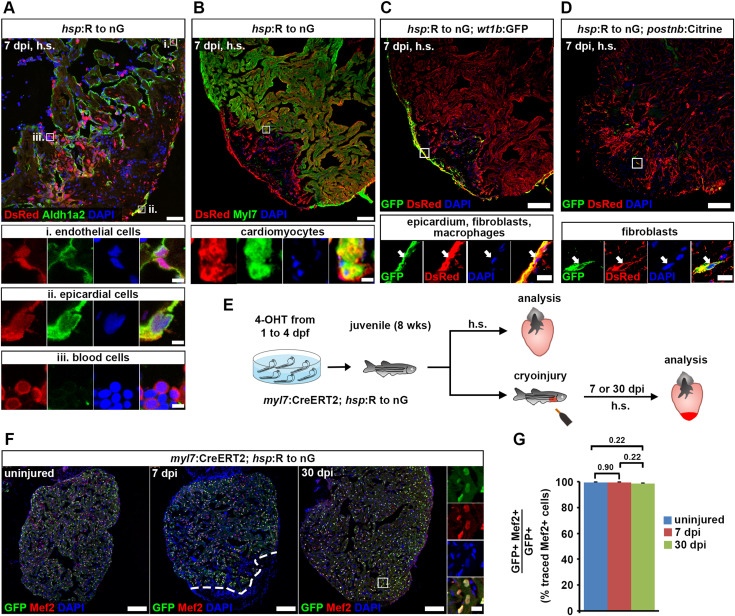
Fig. 7 Fig. 7. Cardiomyocytes remain lineage-restricted during juvenile heart regeneration. (A–D) At 7 dpi in juvenile fish, expression of the hsp:R to nGtud9 tracer transgene is detected by DsRed immunofluorescence after heat-shock (h.s.) in endocardial cells (Aldh1a2+, boxed area i in (A)), epicardial cells (Aldh1a2+, boxed area ii in (A)), blood cells (Aldh1a2— with small, round and intense DAPI-stained nuclei, boxed area iii in (A)), cardiomyocytes (Myl7+, boxed area in (B)), wt1b:EGFPli1Tg+epicardial cells, fibroblasts and macrophages (C), and in postnb:Citrinecn6Tg+fibroblasts (D). Scale bars, 40 μm (overview), 5 μm (magnified view). (E) Experimental design for juvenile cardiomyocyte lineage tracing. (F) Nuclear GFP+ traced cells are widespread throughout the ventricle and colocalize with Mef2, a nuclear cardiomyocyte marker. Scale bars, 100 μm (overview), 12.5 μm (magnified view). (G) No significant difference is detected in average percentages of GFP+ cells expressing Mef2 between uninjured and regenerating hearts. Error bars, s.e.m. n (hearts) = 4 (uninjured), 5 (7 dpi), 6 (30 dpi). n (traced cells) = 858 (uninjured), 823 (7 dpi), 1123 (30 dpi). One-way ANOVA + Holm-Sidak’s multiple comparisons test. Observed difference uninj. vs 7 dpi = 0.06 (0.06% of traced cells in uninj.); smallest significant detectable difference = 0.8 (0.8% of uninj.). Observed difference uninj. vs 30 dpi = 0.8 (0.8% of traced cells in uninj.); smallest significant detectable difference = 1.9 (1.9% of uninj.). Observed difference 7 dpi vs 30 dpi = 0.9 (0.9% of traced cells in 7 dpi); smallest significant detectable difference = 1.6 (1.6% of 7 dpi).
Reprinted from Developmental Biology, 471, Bertozzi, A., Wu, C.C., Nguyen, P.D., Vasudevarao, M.D., Mulaw, M.A., Koopman, C.D., de Boer, T.P., Bakkers, J., Weidinger, G., Is zebrafish heart regeneration "complete"? Lineage-restricted cardiomyocytes proliferate to pre-injury numbers but some fail to differentiate in fibrotic hearts, 106-118, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.