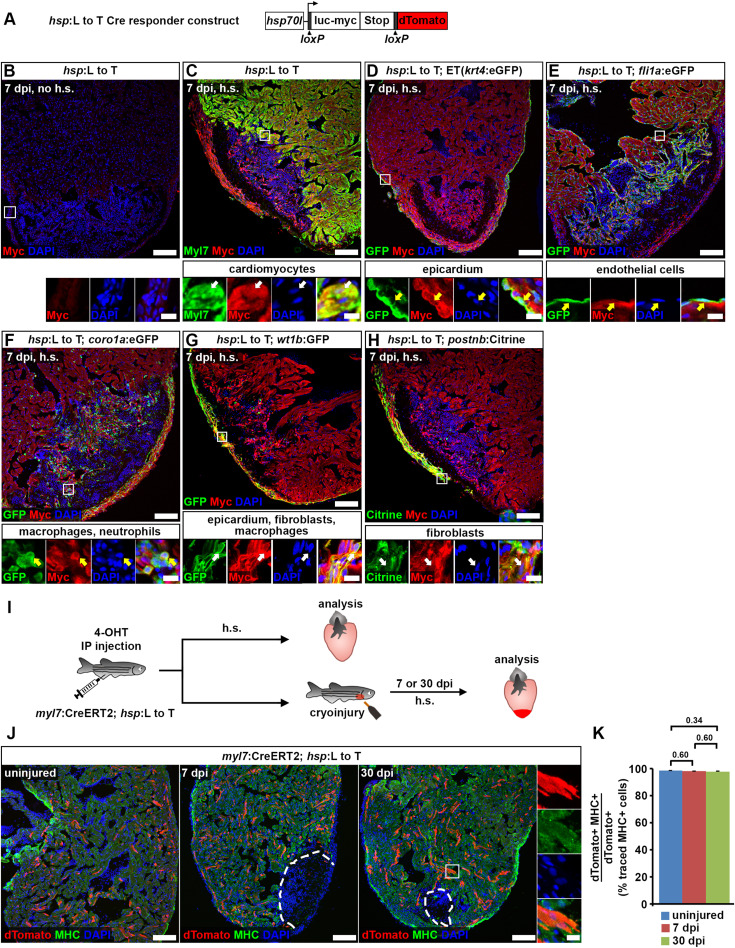
Fig. 5 Fig. 5. Cardiomyocytes remain lineage-restricted during adult heart regeneration. (A) Schematic of the hsp70l:LOXP-luc-myc-STOP-LOXP-dTomato, cryaa:YPetulm9 (hsp:L to T) transgene. (B–H) At 7 dpi, myc-tagged luciferase is not detectable in the ventricle of adult hsp70l:LOXP-luc-myc-STOP-LOXP-dTomato, cryaa:YPetulm9 (hsp:L to T) hearts without heat-shocks (B). After heat-shock, expression is found in myl7+ cardiomyocytes (C), Et(krt4:eGFPsqet27)+ epicardial cells (D), fli1a:eGFPy1Tg+endocardial cells (E), coro1a:eGFPhkz04tTg+leukocytes (F), in wt1b:EGFPli1Tg+epicardial cells, fibroblasts and macrophages (G), and in postnb:Citrinecn6Tg+ activated fibroblasts (H). Boxed areas are shown in magnified view. Scale bars, 100 μm (overview), 10 μm (magnified view). (I) Experimental design for adult cardiomyocyte lineage tracing. (J) dTomato+ traced cells are randomly located throughout the ventricle, and express the cardiomyocyte marker myosin heavy chain (MHC). Scale bars, 100 μm (overview), 12.5 μm (magnified view). (K) No difference is detected in average percentages of dTomato+ traced cells expressing MHC between uninjured and regenerating hearts. Error bars, s.e.m. n (hearts) = 4 (uninjured), 4 (7 dpi), 5 (30 dpi). n (traced cells) = 942 (uninjured), 775 (7 dpi), 1134 (30 dpi). One-way ANOVA + Holm-Sidak’s multiple comparisons test. Observed difference uninj. vs 7 dpi = 0.3 (0.3% of traced cells in uninj.); smallest significant detectable difference = 0.7 (0.7% of uninj.). Observed difference uninj. vs 30 dpi = 0.6 (0.6% of traced cells in uninj.); smallest significant detectable difference = 1.2 (1.2% of uninj.). Observed difference 7 dpi vs 30 dpi = 0.2 (0.2% of traced cells in 7 dpi); smallest significant detectable difference = 1.3 (1.4% of 7 dpi).
Reprinted from Developmental Biology, 471, Bertozzi, A., Wu, C.C., Nguyen, P.D., Vasudevarao, M.D., Mulaw, M.A., Koopman, C.D., de Boer, T.P., Bakkers, J., Weidinger, G., Is zebrafish heart regeneration "complete"? Lineage-restricted cardiomyocytes proliferate to pre-injury numbers but some fail to differentiate in fibrotic hearts, 106-118, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.