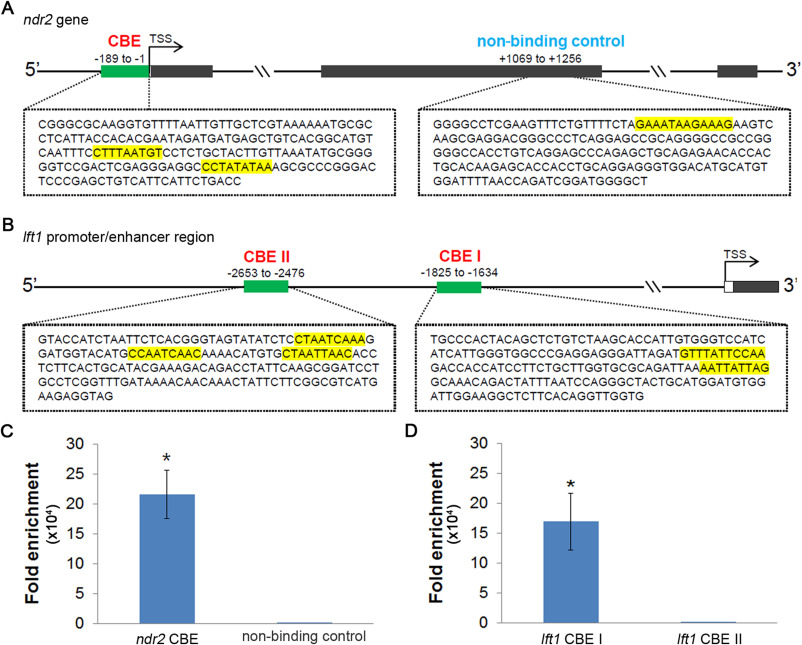
Fig. 6 Fig. 6. Cdx1b directly binds to CDX binding motifs located upstream of ndr2 and lft1 genes. A Genomic structure and nucleotide sequences of CDX binding element (CBE) (−189 to −1 bp) and non-binding control region (+1069 to +1256 bp) of the ndr2 gene are shown. One or two CDX binding motifs located in the exon coding region as non-binding control or CBE are highlighted with yellow. B Genomic structure and nucleotide sequences of CBE I (−1825 to −1634 bp) and CBE II (−2653 to −2476 bp) of the lft1 gene are shown. Two or three CDX binding motifs located in the CBE I or CBE II are indicated by yellow highlighting. C Chromatin DNA from cdx1b-Myc-injected bud embryos was immunoprecipitated using anti-Myc antibody or anti-IgG antibody as a control. The immunoprecipitated DNA was used for qPCR with specific primers spanning the CBE or non-binding control region of ndr2. The CDX binding motifs within CBE but not non-binding control were significantly enriched by anti-Myc antibody compared with IgG control. D The immunoprecipitated chromatin DNA isolated from cdx1b-Myc-injected bud embryos was subjected to qPCR using specific primers spanning CBE I or CBE II of lft1. The CDX motifs within CBE I but not CBE II were significantly enriched by anti-Myc antibody compared with IgG control. Error bars indicate the standard deviation. Student’s t-test was performed. ∗, p < 0.05.
Reprinted from Developmental Biology, 470, Wu, C.S., Lu, Y.F., Liu, Y.H., Huang, C.J., Hwang, S.L., Zebrafish Cdx1b modulates epithalamic asymmetry by regulating ndr2 and lft1 expression, 21-36, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.