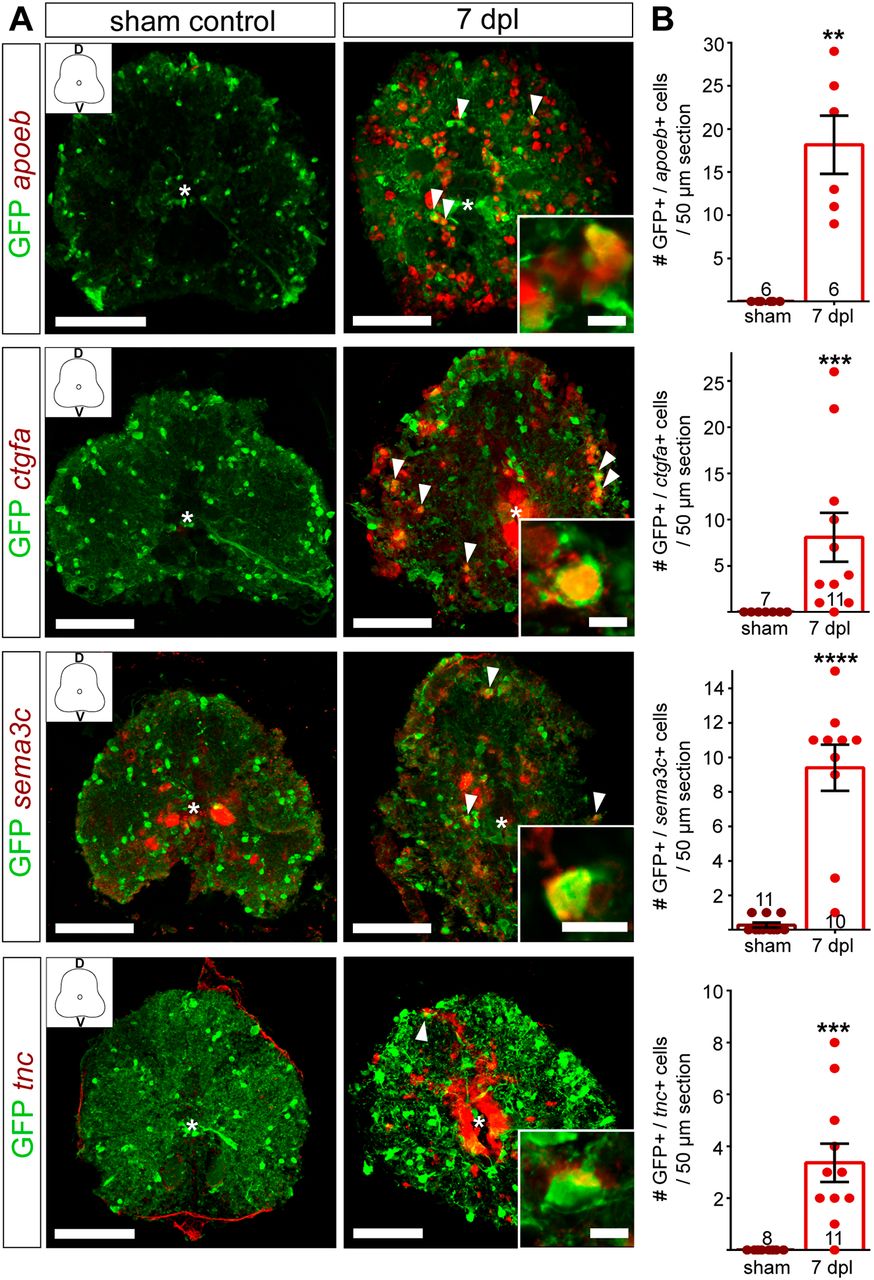
Fig. 8 Validation of RNAseq candidate genes with fluorescent in situ hybridization. (A) Fluorescent in situ hybridization for apoeb, ctgfa, sema3c and tnc in Tg(olig2:eGFP) fish. SCI results in gene expression upregulation (red) in the spinal cord parenchyma (ctgfa, tnc, sema3c and apoeb) and in the central canal (ctgfa and tnc) at 7 dpl. Sections were co-stained using anti-GFP antibody. Overview images are maximum intensity projections of transverse sections. D, dorsal; V, ventral. In the overview images, a white asterisk indicates the central canal. Scale bars: 10 μm in insets; 100 μm in overviews. (B) Quantification of mRNA+/GFP+ cells in 50 µm sections, 350 µm rostral to the lesion site shows a significant increase in the number of OPCs upregulating the respective gene at 7 dpl, compared to sham controls. Data are mean±s.e.m. Numbers in the plots indicate the number of sections counted. Number of experimental animals were as follows: six sham control and six lesioned for apoeb, ctgfa and tnc; eight sham and eight lesioned for sema3c. **P≤0.01; ***P≤0.001; ****P<0.0001 (Mann–Whitney test). Significance is shown compared to sham control.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development