Fig. S1
Defining Imaging Phenotypes, Related to Figure 1, Figure 3, Figure 4
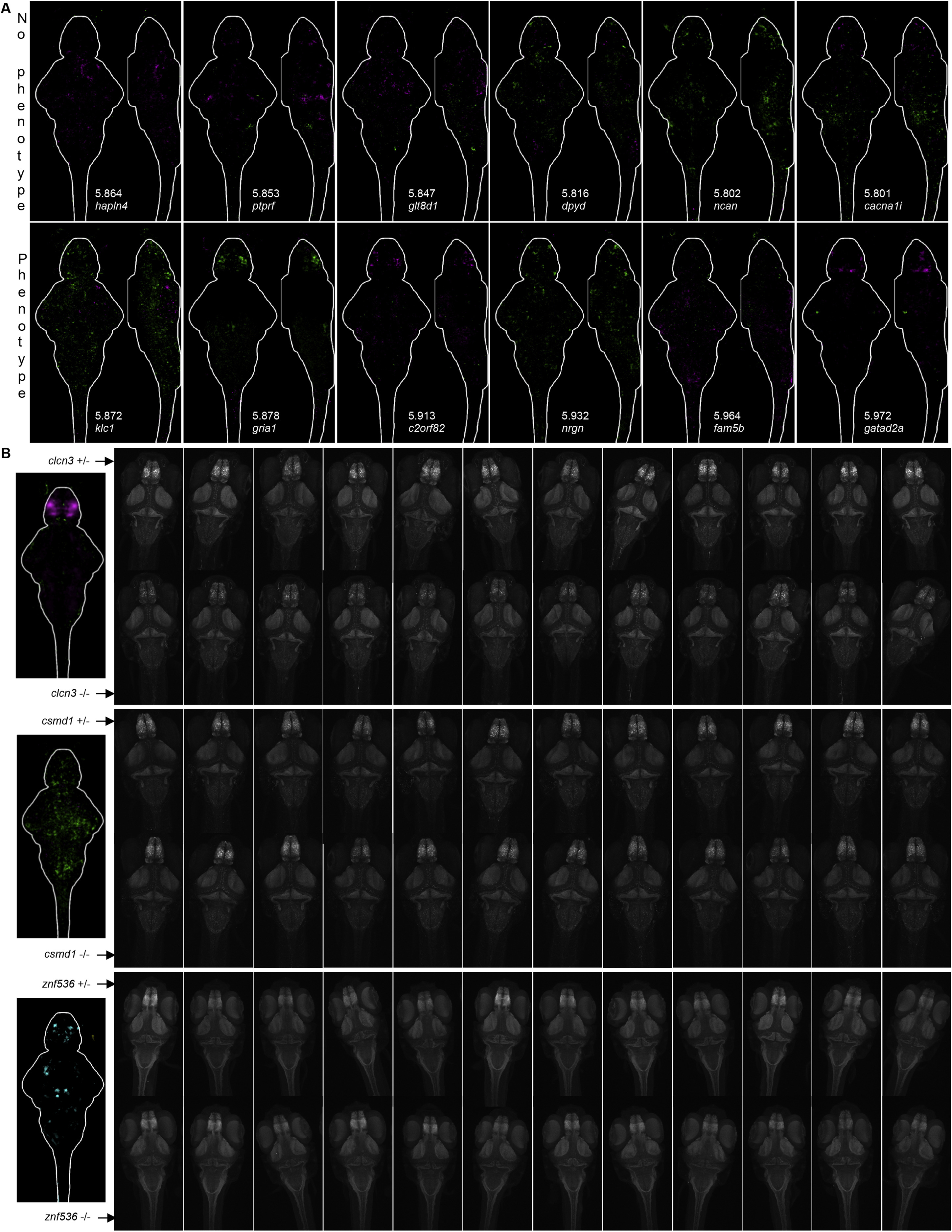
(A) Determination of cutoff for brain activity signals. Images are sum-of-slices intensity projections (Z- and X- axis). This cutoff for the log10(sum of pixels) was made so that mutants with small, specific, repeatable signals were classified as having phenotypes (gria1,forebrain). Full stacks of all individual repeats are available on stackjoint.com/zbrain. Many mutants with diffuse and sparse signals (likely noise) fell below the cutoff. Exceptions included klc1 and nrgn(shown here), which both had signals that reproduced in independent experiments (stackjoint.com/zbrain).
(B) Examples of individual homozygous mutant and heterozygous control brains stained with phospho-Erk (clcn3, csmd1) or total-Erk (znf536) (maximum intensity projections). Twelve brains of each genotype are shown for the three genes. The localized reduction in forebrain activity observed in the clcn3 map is readily observable in the original images. In contrast, the widespread increased activity in the csmd1 map is not as discernable by eye, but is still noticeable in some areas (mesencephalon, rhombencephalon). The MapMAPPING protocol (Randlett et al., 2015) quantifies these less obvious differences. Significant voxels are defined using the Mann-Whitney U statistic Z score, intrinsically selecting for repeatable signals. Variability in brain activity between individuals is most likely due to subtle differences in the experiences of the larvae during the last ten minutes before fixation (the integration time of phospho-Erk). Structural changes in znf536mutants are discernable for the smaller size of the forebrain pallium and symmetric reduction in the cerebellum. The other small signals are likely noise, as they are asymmetric (Figure 3B; znf536 is in the “small changes and noise” group).
Reprinted from Cell, 177(2), Thyme, S.B., Pieper, L.M., Li, E.H., Pandey, S., Wang, Y., Morris, N.S., Sha, C., Choi, J.W., Herrera, K.J., Soucy, E.R., Zimmerman, S., Randlett, O., Greenwood, J., McCarroll, S.A., Schier, A.F., Phenotypic Landscape of Schizophrenia-Associated Genes Defines Candidates and Their Shared Functions, 478-491.e20, Copyright (2019) with permission from Elsevier. Full text @ Cell