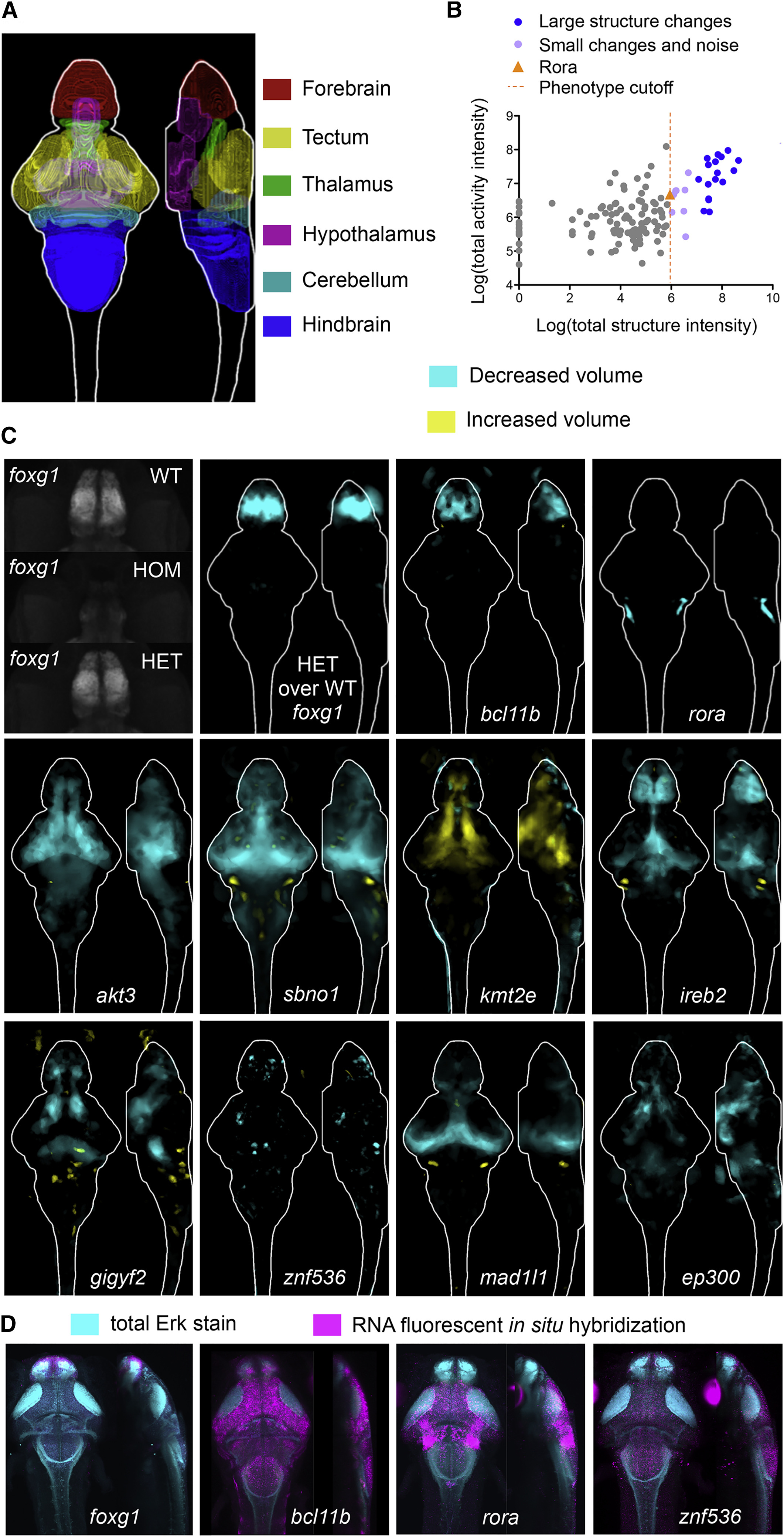
Fig. 3
Whole-Brain Morphology Phenotypes in Zebrafish Mutants
(A) Location of several major regions in the zebrafish brain (Randlett et al., 2015).
(B) Comparison between brain activity and morphology data for all mutants. The rora mutant (orange triangle) represents the smallest (5.95) structural change designated as a phenotype, shown in panel C. Small signals that were not symmetrical were considered noise, compared to small but symmetrical changes as observed in the rora mutant.
(C) Examples of structural differences in mutants calculated using deformation-based morphometry and displayed as sum-of-slices projections (Z- and X- axes). Brain images represent the significant differences in signal between two groups (Randlett et al., 2015), most often homozygous mutants versus heterozygous and/or wild-type siblings (Table S2). The number of animals used in all imaging experiments is available in Table S2 and on stackjoint.com/zbrain(website naming conventions for datasets described in STAR Methods). Brain maps were averaged from two independent clutches of larvae if the experiment was repeated and irreproducible signals were eliminated (STAR Methods). Raw imaging data examples (maximum projections) are shown for the foxg1 mutants, demonstrating that the forebrain in homozygous (HOM) mutants is underdeveloped. A reduction in forebrain size of heterozygous (HET) foxg1 mutants when compared to wild-type (WT) siblings can be quantified with deformation-based morphometry (sum-of-slices projection), although it is not readily apparent in the raw projection.
(D) Fluorescent RNA in situ images for four mutants with altered brain morphology.
Reprinted from Cell, 177(2), Thyme, S.B., Pieper, L.M., Li, E.H., Pandey, S., Wang, Y., Morris, N.S., Sha, C., Choi, J.W., Herrera, K.J., Soucy, E.R., Zimmerman, S., Randlett, O., Greenwood, J., McCarroll, S.A., Schier, A.F., Phenotypic Landscape of Schizophrenia-Associated Genes Defines Candidates and Their Shared Functions, 478-491.e20, Copyright (2019) with permission from Elsevier. Full text @ Cell