Fig. 4
Whole-Brain Activity Phenotypes in Zebrafish Mutants
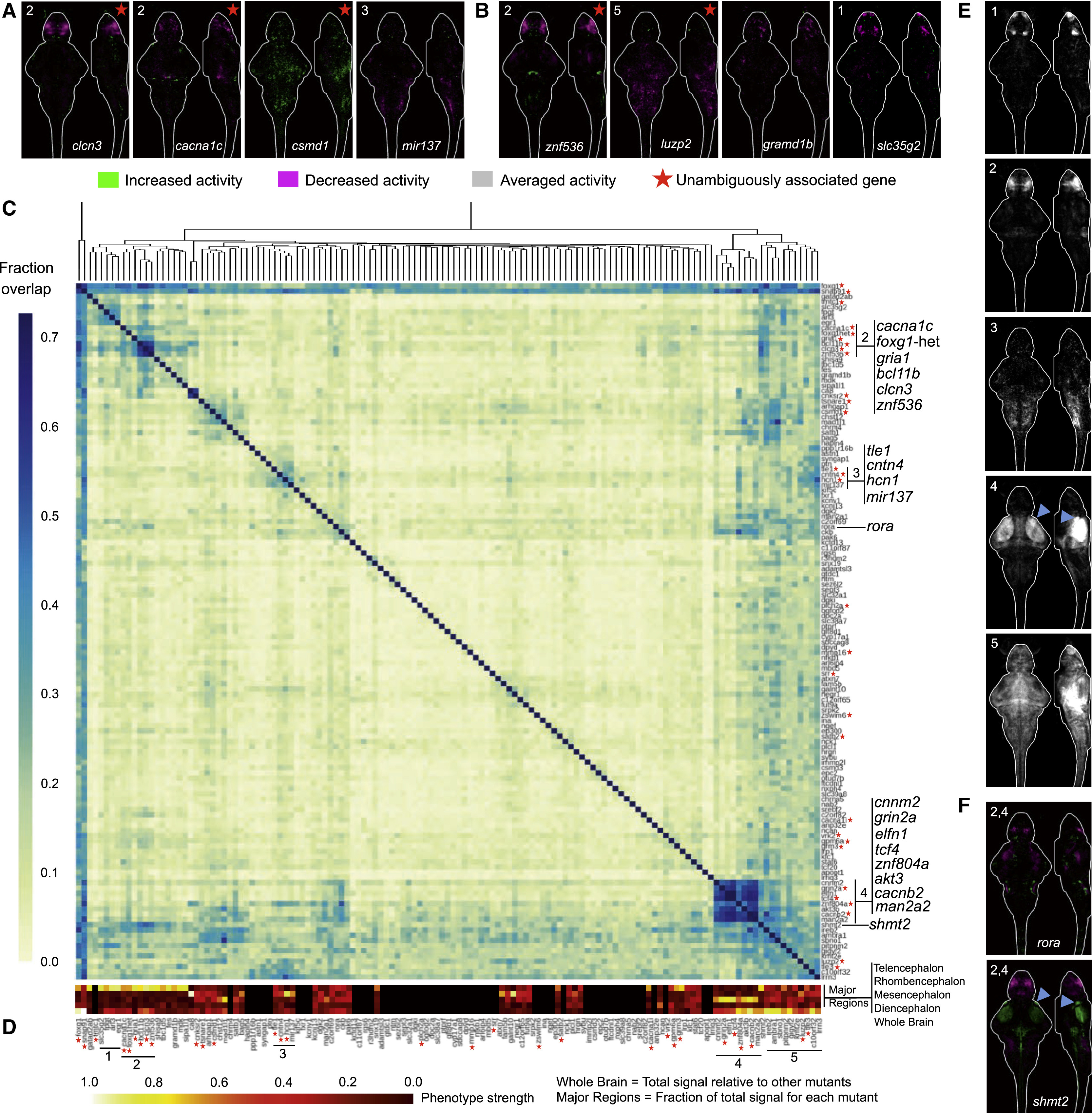
The number of animals used in all imaging experiments is available in Table S2 (average N is 20 for mutant group, 28 for control group) and on stackjoint.com/zbrain (website naming conventions in STAR Methods). Brain maps were averaged from two independent clutches of larvae if the experiment was repeated and irreproducible signals were eliminated (STAR Methods). The white numbers in the upper right corner of images connect pERK activity maps, both of single genes (panels A and B, and Figure S5) and gene averages (panel E), to the heatmaps and to each other. See also Figure S4. See also Figure S5.
(A) Brain activity phenotypes for genes that have been strongly implicated in schizophrenia by previous studies (Table S1). Sum-of-slices projections of significant differences between mutant and control groups of zebrafish larvae (Randlett et al., 2015). The cacna1c mutant phenotype shown is for heterozygous larvae because the homozygous mutant is embryonic lethal (Stainier et al., 1996).
(B) Brain activity phenotypes for four genes that have been minimally studied and have unknown functions.
(C) Percent overlap between mutant brain activity phenotypes was calculated between each image by comparing each brain activity signal to signal in the same location in all other mutant images. These overlaps were then sorted with hierarchical clustering using average linkage. Genes grouped together by this clustering are labeled. The direction of the change in brain activity was disregarded to maximize identification of affected brain regions, and because the direction of the genetic perturbation in human patients is not clear for most genes. The numbers of image stacks that were compared to calculate significant differences in brain activity for each mutant are available in Table S2.
(D) Contribution of each of the four major brain divisions to the overall brain activity phenotype. The signal in each region was divided by the whole brain signal. Prior to dividing the signal in each region by the total signal, the regions were scaled relative to each other based on their respective sizes (rhombencephalon = ∗1, diencephalon = ∗1.76, mesencephalon = ∗1.42, telencephalon = ∗4.36). The original whole brain signal was separately scaled across all mutants with a phenotype, indicating the relevance of the signal in each of the regions. Measures below the cutoff for phenotype designation (Figure S1, Figure S2) are displayed in black.
(E). Average signal for mutants with similar brain activity maps. Examples of individual maps that are included in these averages are labeled in panels A and B, as well as in Figure S5. The blue arrow highlights retinal arborization field AF7.
(F) Two mutants with brain activity signals that overlap with both the forebrain (group 2) and the tectum (group 4).
Reprinted from Cell, 177(2), Thyme, S.B., Pieper, L.M., Li, E.H., Pandey, S., Wang, Y., Morris, N.S., Sha, C., Choi, J.W., Herrera, K.J., Soucy, E.R., Zimmerman, S., Randlett, O., Greenwood, J., McCarroll, S.A., Schier, A.F., Phenotypic Landscape of Schizophrenia-Associated Genes Defines Candidates and Their Shared Functions, 478-491.e20, Copyright (2019) with permission from Elsevier. Full text @ Cell