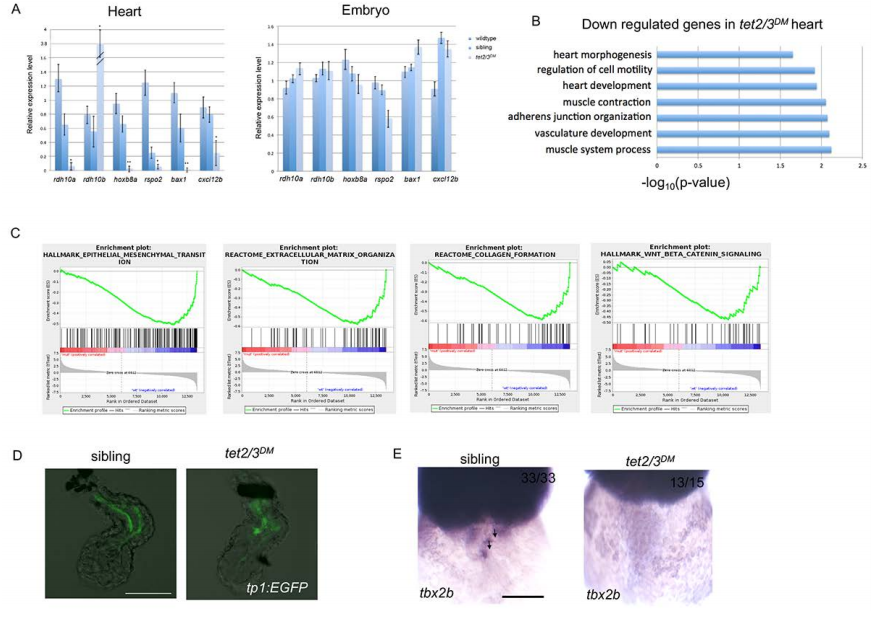
Fig. S4
tet2/3DM Larvae Show Cardiac AVC Defects at 48-hpf. Related to Figure 3 and Figure 4.
(A) Quantitative RT-PCR validation of top down-regulated genes from RNA sequencing. RT-PCR analysis ofrdh10a, hoxb8a, rspo2, bax1, cxcl12b transcripts in 2-dpf embryonic heart or whole embryo. Notably, rdh10bapparently compensates for the rdh10a loss, with 4-fold increase in tet2/3DM hearts, which is consistent with RNA sequencing results.
(B) GO analysis shows down-regulated biological pathways in 48 hpf tet2/3DM hearts compared to wildtype hearts by RNA sequencing using isolated hearts.
(C) Gene set enrichment analysis shows down-regulated biological pathways in 48-hpf tet2/3DM hearts compared to wildtype hearts by RNA sequencing using isolated hearts.
(D) Normal Notch Activity in tet2/3DM endocardium. GFP-labeled atrial endocardium indicates Notch activity in sibling hearts as well as tet2/3DM hearts. Hearts were dissected from 48 hpf larvae carrying the Tg(tp1:EGFP) Notch reporter transgene.
(E) WISH for AVC markers tbx2b at 48 hpf. Black arrows indicate AVC specific expression of tbx2b in sibling but not tet2/3DM heart. Scale bars: 50μm.