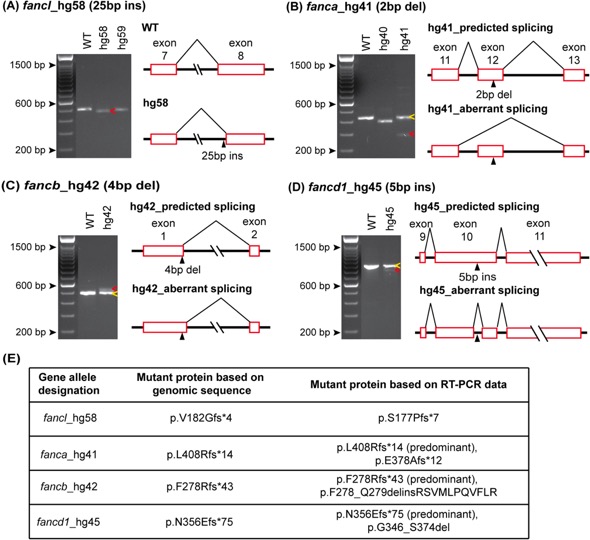
Fig. 1 RT-PCR confirms indel mutations and identifies aberrant splice variants caused by CRISPR/Cas9 mutation.
(A-D) WT and mutant allele RT-PCR products resolved on 2% agarose gel are shown in the left panels and splicing aberrations are depicted in the right panels. (A) The observed RT-PCR product for fancl_hg58 mutant was smaller in size (red arrow) than expected based on the 25 bp insertion in exon 8. The sequence of the hg58 product showed shift in its splice acceptor site resulting in loss of 48 bp sequence, that includes 23 bp from exon 8 beginning and CRISPR/Cas9 induced 25 bp insert. (B-D) Partial activation of cryptic splice site near the indel mutation was observed in fanca_hg41, fancb_hg42 and fancd1_hg45 mutant alleles. The RT-PCR products for these mutants showed two bands, one matched expected size (yellow arrows) and the other was from an altered splice product (red arrows). PCR products were cloned and sequenced to determine aberrant splice product sequence. In hg41 mutant, the indel mutation (2 bp del) in exon 12 results in partial skipping of mutated exon (B). In hg42 mutant, the indel mutation (4 bp del) in exon 1 results in partial use of new splice donor site in intron 1 (C). In hg45 mutant, the indel mutation (5 bp ins) in exon 10 results in partial use of new splice acceptor and donor in exon 10 (D). (E) The consequence on the predicted encoded protein based on the genomic indel mutation, and the observed aberrant splice product for all four mutant lines are shown. RT-PCR gel image data for all 36 alleles is shown in S3A Fig.