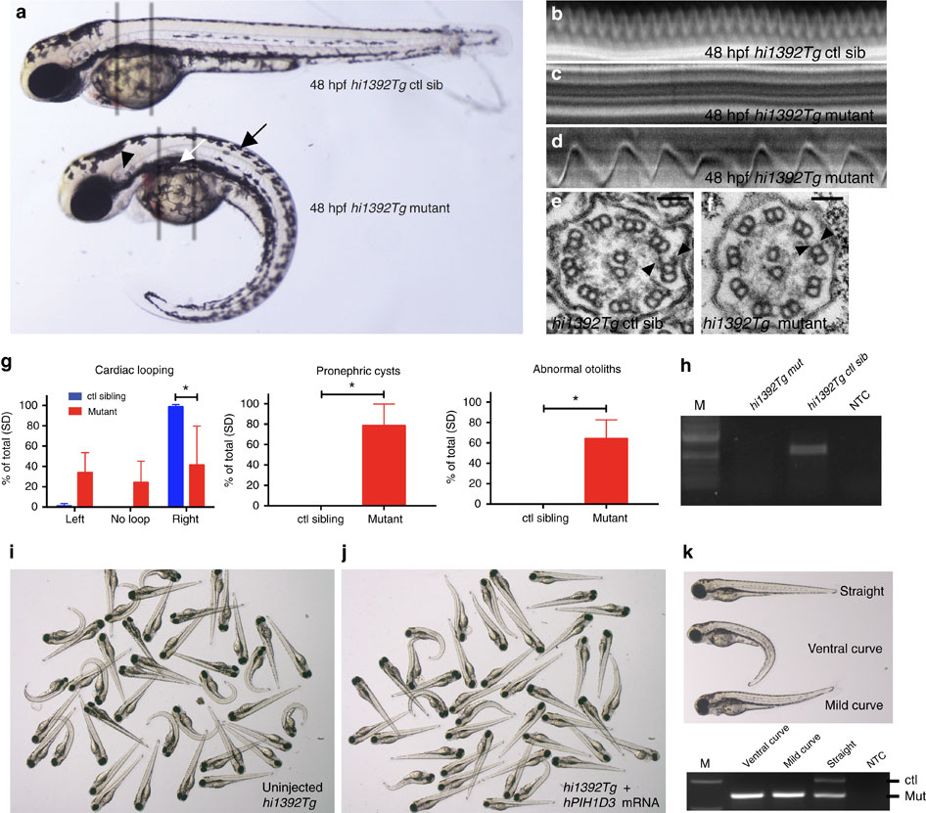
Fig. 4
(a) Comparison of a representative hi1392Tg control sibling (ctl sib, top), and a hi1392Tg mutant (bottom) harbouring a transgenic insertion in the pih1d3 (previously known as twister) gene, at 48 h post fertilization (48 hpf). Mutants display an abnormally curved body axis (black arrow), pronephric cysts (white arrow), abnormal otolith formation (black arrowhead). (b–f) Cilia defects are evident in hi1392Tg mutants at 48 hpf, with the grey bars in a highlighting areas used for pronephric cilia imaging shown in b–f. Cilia motility defects are evident in representative kymographs recorded in hi1392Tg mutant siblings (c,d) compared with hi1392Tg control siblings (b), created from 0.5-s, 1,000 fps videos of single pronephric cilia bundles beating in 48 hpf (representative of 16 recordings). Mutants had mainly static cilia (c) with an occasional cilia showing retained reduced beating (d). Also see Supplementary Videos 6,7,8,9. (e,f) TEM of 48 hpf pronephric cilia cross-sections from the same area reveal a reduction and loss of outer and IDAs (arrowheads) in hi1392Tg mutants (f) compared with control siblings where the arms are visible (e). Scale bar, 50 nm. (g) Quantification of common phenotypes of hi1392Tg mutant embryos (red columns) compared with control sibling embryos (blue columns). hi1392Tg mutant embryos display 64–84% levels of abnormal (that is, absent or reverse) cardiac looping, pronephric cysts and otolith malformations, while this was never, or rarely, seen in control siblings. Otolith malformation was recorded if there were more or less than two observed. Columns represent data from three separate experiments assessing phenotype in 48 hpf embryos (n=40–71 per experiment). Error bars show standard deviation with t-test, P-value *<0.05. (h) The insertion hi1392Tg creates a null allele: RT-PCR of a pool of 15 embryos indicates a complete lack of pih1d3 transcript expression in hi1392Tg mutants compared with a pool of 15 control siblings. (i–k) Examples of clutches of 72 hpf embryos from hi1392Tg/+ crosses injected at one-cell stage with 10 pg of human PIH1D3 mRNA show a reduction in mutant phenotypes (j) compared with non-injected hi1392Tg control siblings (i), with representative embryos with a straight, ventral curve, or mild curve phenotype shown in k, upper panel. PCR genotyping in all three of these groups shows the lower hi1392Tg insertion allele band present, but the upper control allele band is present only in the straight-tailed siblings (k, lower panel). M, DNA size marker; NTC, non-template control.