Fig. 5
Ttc28 Is Required to Regulate Microtubule Dynamics
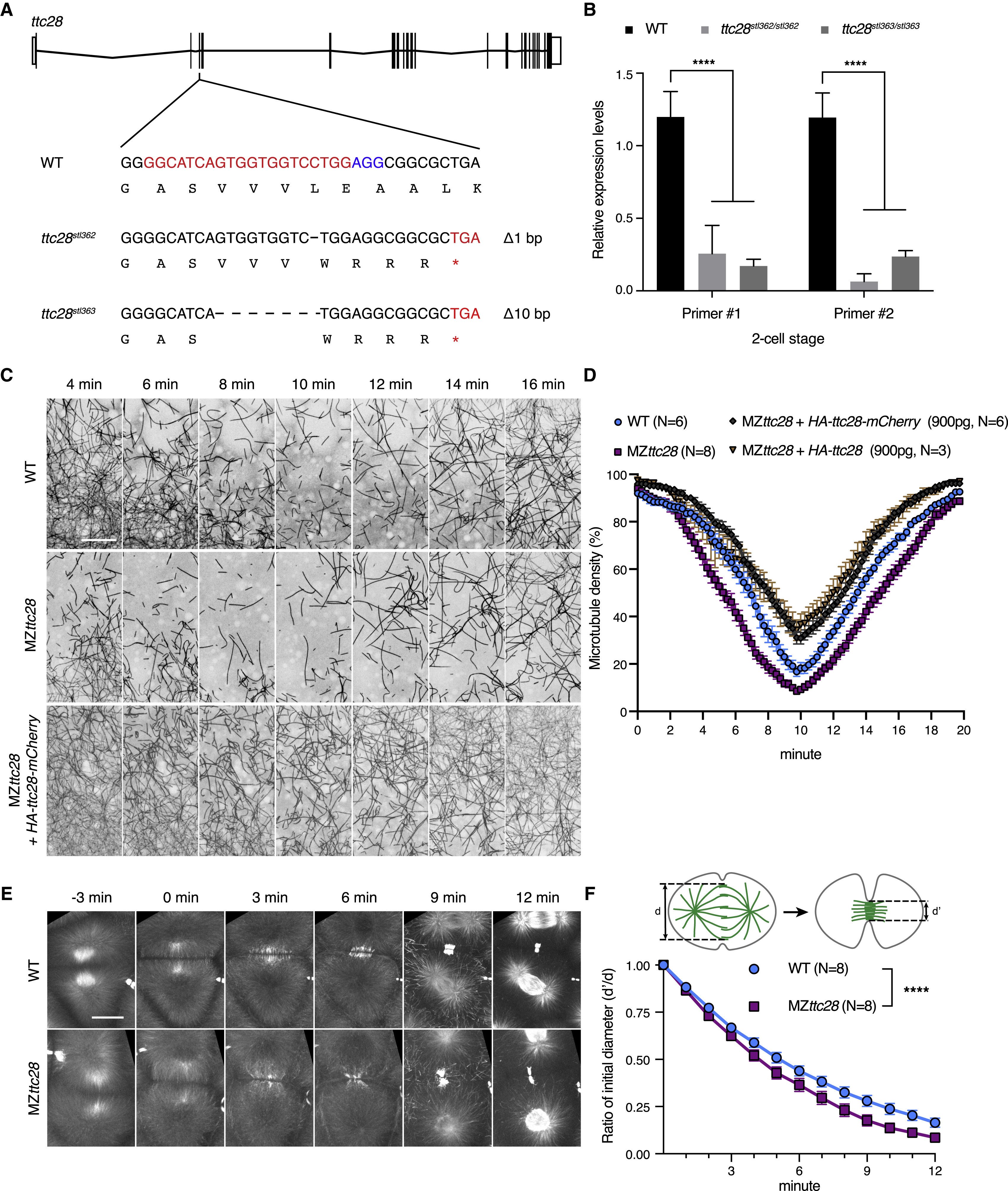
(A) Schematic of ttc28 single guide RNA targeting site and the sequences of ttc28stl362 and ttc28stl363 mutant alleles. Asterisk indicates premature stop codon.
(B) qPCR analyses of ttc28 transcript levels in WT, MZttc28stl362/stl362, and MZttc28stl363/stl363 mutants at two-cell stage. Two different sets of primers were used and normalized to β-actin. ∗∗∗∗p < 0.0001, Student’s t test. Error bars represent SD. N = 3 biological repeats.
(C) Representative time-lapse still images of YCL microtubules at 1.75–2.5 hpf in WT, MZttc28stl363/stl363, and HA-Ttc28-mCherry RNA-injected (900 pg) MZttc28stl363/stl363Tg[ef1α:dclk-GFP] embryos. Scale bar, 30 μm.
(D) Quantification of YCL microtubule density from time-lapse videos in WT, MZttc28stl363/stl363, and HA-Ttc28 or HA-Ttc28-mCherry RNA-injected (900 pg) MZttc28stl363/stl363 embryos at 1.75–2.5 hpf. Error bars represent SEM. N, number of embryos.
(E) Representative time-lapse still images of microtubule dynamics during cytokinesis in WT and MZttc28stl363/stl363 embryos using Tg[ef1α:dclk-GFP]. Scale bar, 30 μm.
(F) Quantification of midzone microtubule assembly rates in WT and MZttc28stl363/stl363 blastulae at 1.75–2.5 hpf. ∗∗∗∗p < 0.0001, paired Student's two-tailed t test. Error bars represent SEM. N, number of embryos.
See also Figures S5 and S6, Videos S6 and S7.
Reprinted from Developmental Cell, 45, Chen, J., Castelvecchi, G.D., Li-Villarreal, N., Raught, B., Krezel, A.M., McNeill, H., Solnica-Krezel, L., Atypical Cadherin Dachsous1b Interacts with Ttc28 and Aurora B to Control Microtubule Dynamics in Embryonic Cleavages, 376-391.e5, Copyright (2018) with permission from Elsevier. Full text @ Dev. Cell