Fig. s1
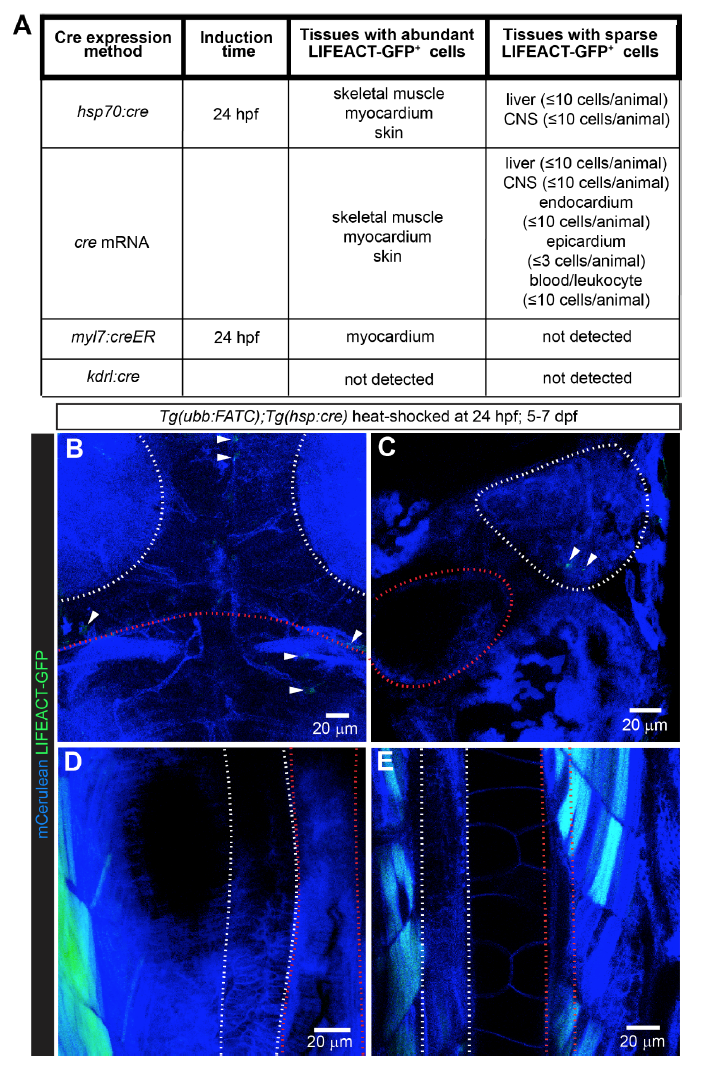
FATC reporter expression is restricted to a few tissues. A, List of tissues containing LIFEACT-GFP+ cells, observed between 4-7 dpf, upon ubiquitous and cell-type specific cre expression in Tg(ubb:FATC) animals. Ubiquitous cre expression by the heat-inducible Tg(hsp70:cre) or injection of cre mRNA in Tg(ubb:FATC) embryos gave rise to LIFEACT-GFP+ skeletal and cardiac myocytes, and skin cells. A few cells in the liver and central nervous system (CNS), were also LIFEACT-GFP+. 4-OHT induction of cardiomyocyte-specific cre expression in Tg(ubb:FATC);Tg(myl7:creER) embryos at 24 hpf led to LIFEACT-GFP+ cardiomyocytes. No LIFEACT-GFP+ cells were detectable in Tg(ubb:FATC);Tg(kdrl:cre) animals in which cre is constitutively expressed in endothelial lineage cells. B-E, Embryos from Tg(ubb:FATC) and Tg(hsp:cre) crosses were heat-shocked at 24 hpf to induce ubiquitous cre expression. While the expression of the ubb:FATC transgene was observed by membrane Cerulean fluorescence (blue) in all tissues, a few LIFEACT-GFP+ cells (green, arrowheads) were detectable in the CNS (B) and the liver (C) at 5-7 dpf. The muscle (D and E) and the skin (not shown) were the only tissues that contained significant numbers of LIFEACT-GFP+ cells. Areas enclosed by white dashed lines are the tectal neuropil (B), liver (C), pancreas (D), and spinal cord (E). Red dashed lines mark the midbrain-hindbrain boundary (B), gall bladder (C), intestine (D), and dorsal aorta (E). All images are maximum intensity projections of 20-60 μm thick confocal stacks. B is dorsal view, anterior up. C-E are lateral views, dorsal to the left, anterior up. 12-39 animals were examined for each cre expression method.