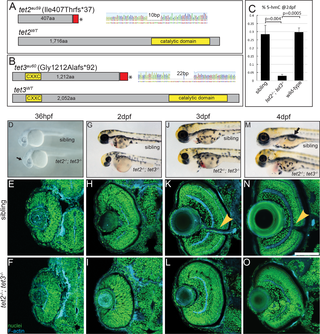
Fig. 1
(A) tet2au59 mutants possess a 10bp deletion resulting in a frameshift, insertion of 37 incorrect aa’s (red), and a premature stop codon, truncating the protein at amino acid (aa) 444 of 1,716aa. (B) tet3au60 mutants possess a 22bp deletion, resulting in a frameshift, insertion of 92 incorrect aa’s (red), and a premature stop codon, truncating the protein at aa 1,304 of 2,052. (C) Genomic DNA isolated from 2dpf tet2-/-;tet3-/- mutants shows a >9 fold reduction in global 5hmC levels when compared to phenotypically wild-type siblings and genetically wild-type embryos in an ELISA (n = 20 embryos, p = 0.004 and p = 0.0005, respectively; Error bars = ± 1 S.D.). (D) At 36hpf, tet2-/-;tet3-/- mutants are identifiable based on a kinked head and slightly enlarged brain (arrow). (E,F) Retinae of 36hpf tet2-/-;tet3-/- mutants are morphologically similar to wild-types. (G) At 2dpf, tet2-/-;tet3-/- mutants are microphthalmic, possess cardiac edema and their heads are smaller than phenotypically wildtype siblings. (H-I) The retina is not laminated, with retinal cells appearing progenitor-like in morphology when compared to phenotypically wild-type siblings. (J) Cardiac edema becomes progressively enlarged at 3dpf in tet2-/-; tet3-/- embryos. (K,L) The retina remains poorly laminated in tet2-/-;tet3-/- mutants, and they lack a morphologically obvious optic nerve (arrowhead in sibling). (M) A 4dpf tet2-/-; tet3-/- embryos do not possess an inflated swim bladder (arrow). (N,O) The retina remains poorly laminated and they lack a morphologically obvious optic nerve (arrowhead in sibling). DNA (green), F-actin (cyan). Dorsal is up and anterior to the left. Scale bar = 80μm.