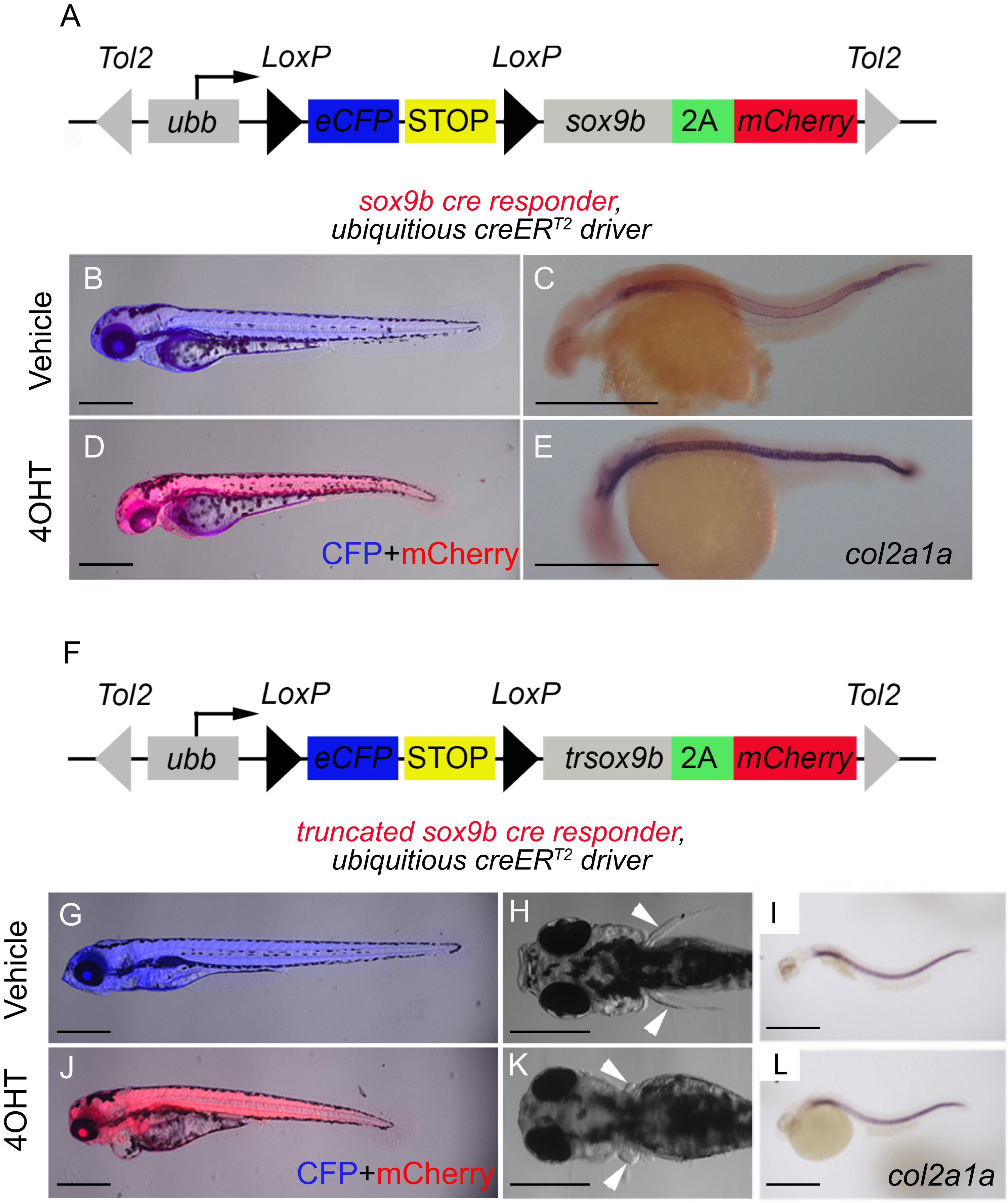
Fig. 3
Inducible systems to overexpress Sox9b or dominant-negative Sox9b. (A) sox9b and (F) trsox9b cre responder constructs used to make new lines. Tol2 arms (gray triangles) facilitate transgenesis. Following cre-recombination between LoxP sites (black triangles), both the eCFP gene (blue) and stop sequences (yellow) are removed allowing ubiquitous expression of either sox9b-2A-mCherry (A) or trsox9b-2A-mCherry (F) to be driven off the ubb promoter/enhancer. Due to 2A sequence (green), either Sox9b (A) or trSox9b (F) will be expressed as separate proteins along with mCherry. Double transgenic larvae for the ubiquitous creERT2driver and either sox9b cre-responder (B-E) or trsox9b cre-responder (G-J). (B, D, G, J) Merged fluorescence images of the larvae at 72 hpf to detect both eCFP in blue and mCherry in red. (C, E, I, L) Results of whole mount in situ hybridization on 24 hpf larvae to detect the known Sox9 target gene col2a1a. (H, K) Bright field dorsal images at 72 hpf. From 4 hpf, fish in (B, C, G-I) were treated with vehicle and fish in (D, E, J-L) were treated with 5 ¼M 4OHT. 4OHT treatment leads to induction of mCherry expression (D and J) concomitant with abnormal gross development due to Sox9b overexpression (D) and trSox9b expression (J-K). Overexpression of Sox9b also caused increased expression of the Sox9 target gene col2a1a (E). Scale bars=500 µm.
Reprinted from Developmental Biology, 418(1), Huang, W., Beer, R.L., Delaspre, F., Wang, G., Edelman, H.E., Park, H., Azuma, M., Parsons, M.J., Sox9b is a mediator of retinoic acid signaling restricting endocrine progenitor differentiation, 28-39, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.