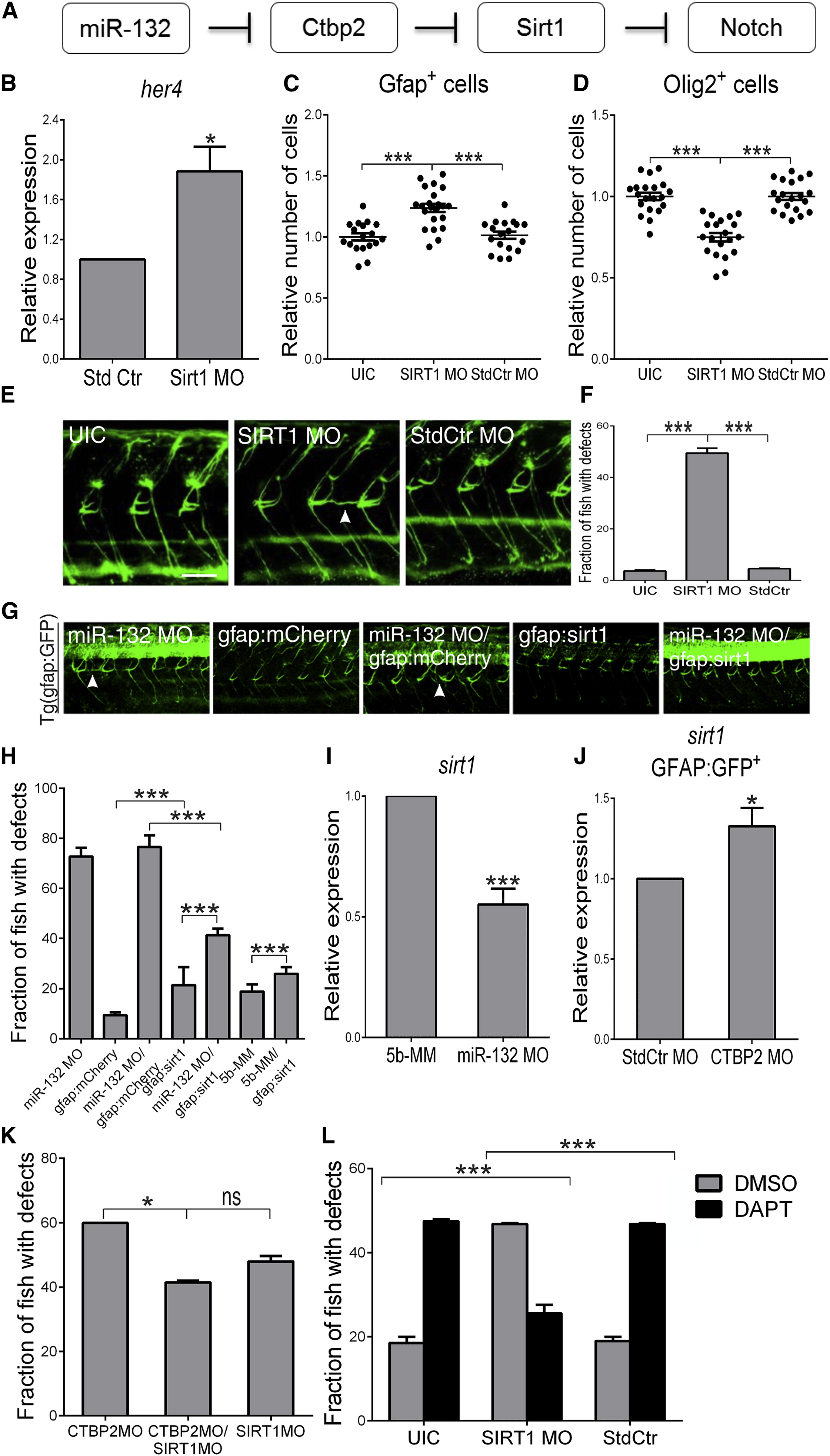
Fig. 5
The Histone Deacetylase Sirt1 Is Regulated by miR-132 and Ctbp2
(A) Model of Sirt1 involvement in the miR-132/Ctbp2/Notch regulatory pathway.
(B) Semiquantitative PCR analysis of her4 expression levels in Sirt1 morphants (SIRT1 MO) and control embryos (Std Ctr) at 24 hpf. Values were normalized as described earlier.
(C and D) Effect of Sirt1 downregulation (SIRT1 MO) on the total number of Gfap+ (C) and Olig2+ (D) in the spinal cord of Sirt1 morphant and control Tg(gfap:GFP) (C) or Tg(olig2:GFP) (D) embryos at 48 hpf. Each dot of the scatterplot represents normalized averages as described earlier.
(E) Whole-mount immunostaining with zrf-1 at 72 hpf, lateral views of trunk. SIRT1 MO, Sirt1 morphants; StdCtr MO, larvae injected with standard control morpholino. Scale bar, 50 µm.
(F) Quantification of glial arborization phenotype in Sirt1 morphants at 72 hpf. Sample size, n = 75 for UIC; n = 90 for SIRT1 MO; n = 95 for StdCtr MO.
(G) Lateral trunk views of 72 hpf - larvae injected with miR-132 morpholino (miR-132 MO), gfap:mCherry (mCherry under the gfap promoter), gfap:sirt1 (sirt1 under the gfap promoter) or coinjected with miR132 morpholino and gfap:mCHERRY or miR-132 morpholino and gfap:sirt1. Scale bar, 50 µm. In (E) and (G), arrowheads indicate ectopic Gfap+ glial processes.
(H) Quantification of the partial rescue of the glial arborization phenotype in the miR-132 morphants overexpressing Sirt1 under the gfap promoter at 72 hpf. Sample sizes: n = 52 for miR-132 MO; n = 61 for gfap:mCherry; n = 57 for miR-132 MO/gfap:mCherry; n = 63 for gfap:sirt1; n = 72 for miR-132 MO/gfap:sirt1; n = 70 for 5b-MM; and n = 58 for 5b-MM/gfap:sirt1.
(I and J) Semiquantitative real-time PCR analysis showing expression of sirt1 in miR-132 morphant (miR-132 MO) and control embryos (5b-MM) at 24 hpf (I) or in gfap:GFP+ FACS-sorted cells isolated from Tg(gfap:GFP) Ctbp2 morphants compared to control larvae, at 72 hpf (J). Values were normalized as described earlier.
(K) Quantification of glial arborization phenotype in Ctbp2/Sirt1 double morphants. Sample sizes: n = 78 for CTBP2 MO (translation blocking morpholino); n = 95 for CTBP2 MO/SIRT1 MO; and n = 87 for SIRT1 MO.
(L) Quantification of the glial arborization phenotype at 72 hpf in UIC, Sirt1 morphants (SIRT1 MO), and larvae injected with a standard control morpholino (StdCtr) following 48 hr incubation in DAPT or vehicle control (DMSO). Sample sizes: n = 87 for UIC-DAPT; n = 74 for UIC-DMSO; n = 66 for SIRT1 MO-DAPT; n = 69 for SIRT1 MO-DMSO; n = 54 for StdCtr-DAPT; and n = 66 for StdCtr-DMSO. Values are presented as mean ± SEM. *p < 0.05; ***p < 0.001; ns, nonsignificant.
Reprinted from Developmental Cell, 30(4), Salta, E., Lau, P., Sala Frigerio, C., Coolen, M., Bally-Cuif, L., De Strooper, B., A Self-Organizing miR-132/Ctbp2 Circuit Regulates Bimodal Notch Signals and Glial Progenitor Fate Choice during Spinal Cord Maturation, 423-36, Copyright (2014) with permission from Elsevier. Full text @ Dev. Cell