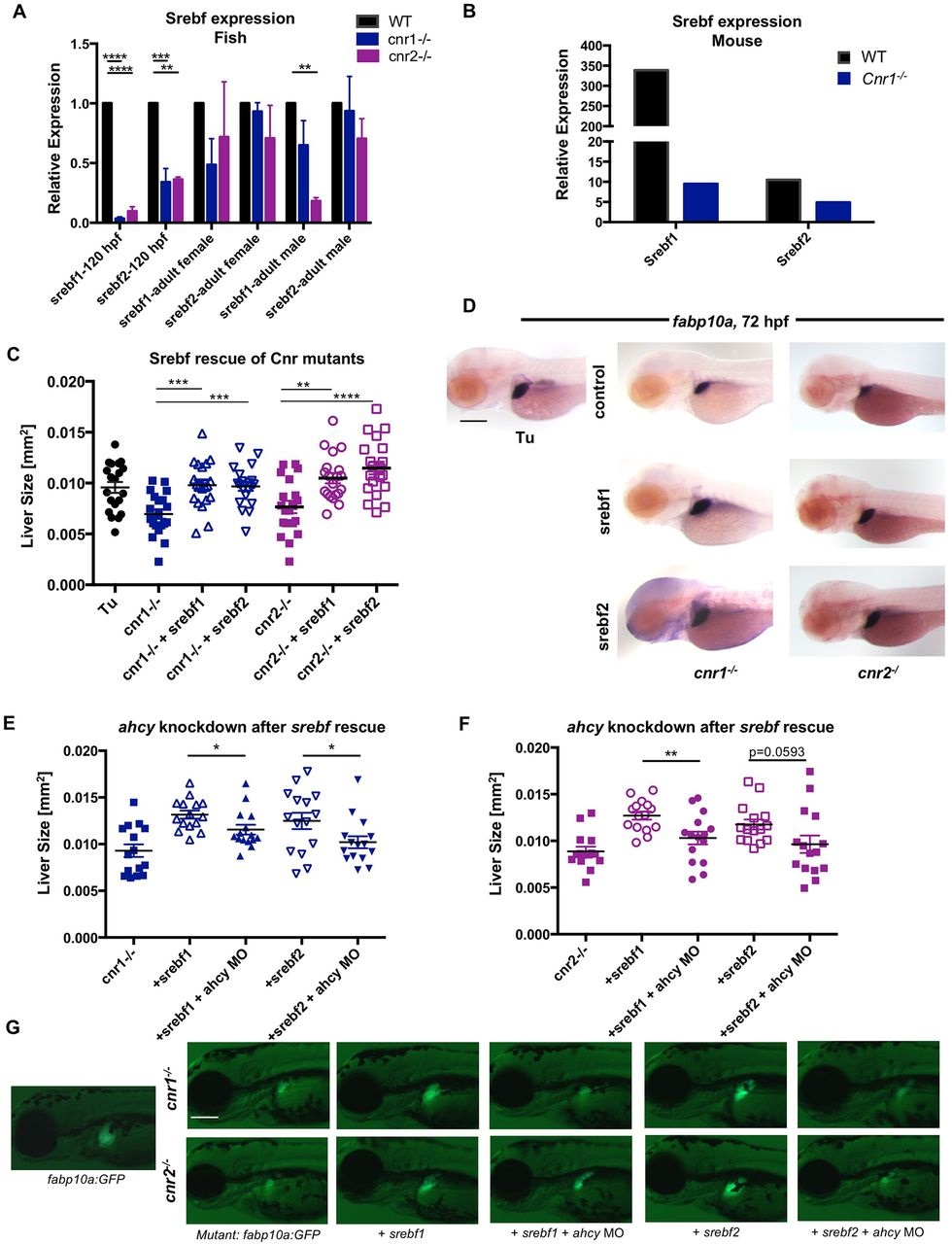
Fig. 7
EC signaling regulates methionine metabolism via Srebfs in Cnr mutants. (A) srebf1 and srebf2 expression in pooled WT, cnr1-/- and cnr2-/- zebrafish embryos at 120hpf, and in adult livers, quantified by qRT-PCR. Data analyzed using the ΔΔCt method. Mean±s.e.m.; n=3 pooled biological samples of 20 embryos each. One-way ANOVA. ****P<0.0001, ***P<0.001 and **P<0.01 for srebf1 or srebf2 expression in mutants versus WT. (B) Srebf1 and Srebf2 expression in male Cnr1-/- mice. n=6. (C,D) Scatter plot of liver morphometric measurements (C) and fabp10a ISH (D) at 72hpf in cnr1-/- and cnr2-/- mutants with and without overexpression of srebf1 and srebf2 mRNA. Results shown represent one independent experiment of triplicate experiments which yielded similar results. Mean±s.e.m.; n>15. One-way ANOVA. ***P<0.001 for cnr1-/- control versus srebf1 or srebf2 overexpression; **P<0.01 for cnr2-/- control versus srebf1 overexpression; ****P<0.0001 for cnr2-/- control versus srebf2 overexpression. (E-G) Scatter plot of liver morphometric measurements (E,F) and live imaging (G) of cnr1-/-; fabp10a:GFP and cnr2-/-; fabp10a:GFP at 72hpf with overexpression of srebf1 and srebf2 in the presence and absence of concurrent ahcy morpholino knockdown. Results represent one independent experiment of triplicate experiments yielding similar results. Mean±s.e.m.; n>15. Student′s t-test. *P<0.05 in cnr1-/- background (E); **P<0.01 in cnr2-/- background (F). Scale bars: 0.2mm.