Fig. 1
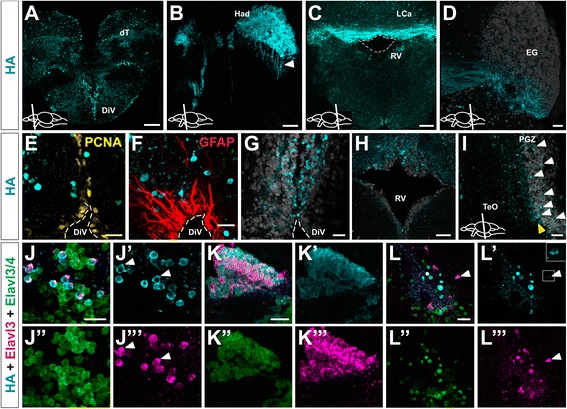
Distribution of HA-Uprt-mCherry transgene expression on cross sections of adult zebrafish brain. a Section showing HA labeling (cyan) in the dorsal telencephalon (dT) and the diencephalic ventricle (DiV). b High magnification of the dorsal habenula (Had) in the anterior diencephalon, with HA labeling in cells and projections (white arrowhead) from the Had to the interpeduncular nucleus (IPN) in the mesencephalon. c Section through the cerebellum showing HA-localization near the rhombencephalic ventricle (RV, Lobus caudalis cerebelli (LCa)). d HA-expression in the ventral half of the eminentia granularis (EG) and axon projections. Elavl3/4 labeling (white) marks most cells of the EG. e Co-labeling of HA and the proliferation marker PCNA (yellow) in a high magnification of the diencephalic ventricle (DiV) shows no overlap. f HA-labeling and the glial marker GFAP (red) are not co-expressed in the same cells. g HA-positive cells in the vicinity of the diencephalic ventricle. Elavl3/4 labeling (white) partially overlaps with HA labeling (cyan) but shows a much broader expression domain than HA labeling. h Most HA-positive neurons are located in the dorsal part of the rhombencephalic ventricle (RV). The ventricle is outlined by Elavl3/4 labeling (white). i Some HA-positive neurons are located within the periventricular gray zone (PGZ, white arrowheads). Many HA-labeled neurons are located at the margin of the PGZ and the optic tectum (TeO, yellow arrowhead). Elavl3/4 labeling marks most cells of the PGZ. j High magnification of the diencephalic ventricular zone showing co-expression of HA (cyan) and endogenous Elavl3 (magenta) within the population of Elavl3/4-positive neurons (green). k Overlapping expression of HA, Elavl3 and Elavl3/4 in the dorsal habenula. l Ventrally located nucleus in the hypothalamus shows that all Elavl3-positive neurons co-express HA-Uprt-mCherry (white arrowhead and inset). Cartoons in a-d and i show lateral views of adult zebrafish brain indicating the level of the individual cross sections. e-g and i. Sections show a similar level as in a or c, respectively. The panels j′, j′′, j′′′, k′, k′′, k′′′ and l′, l′′, l′′′ show single channels. Scale bars represent 100 µm in a; 40 µm in b, g; 150 µm in c; 45 µm in d, h; 25 µm in e, f; 35 µm in i; 20 µm in h, k; and 30 µm in l