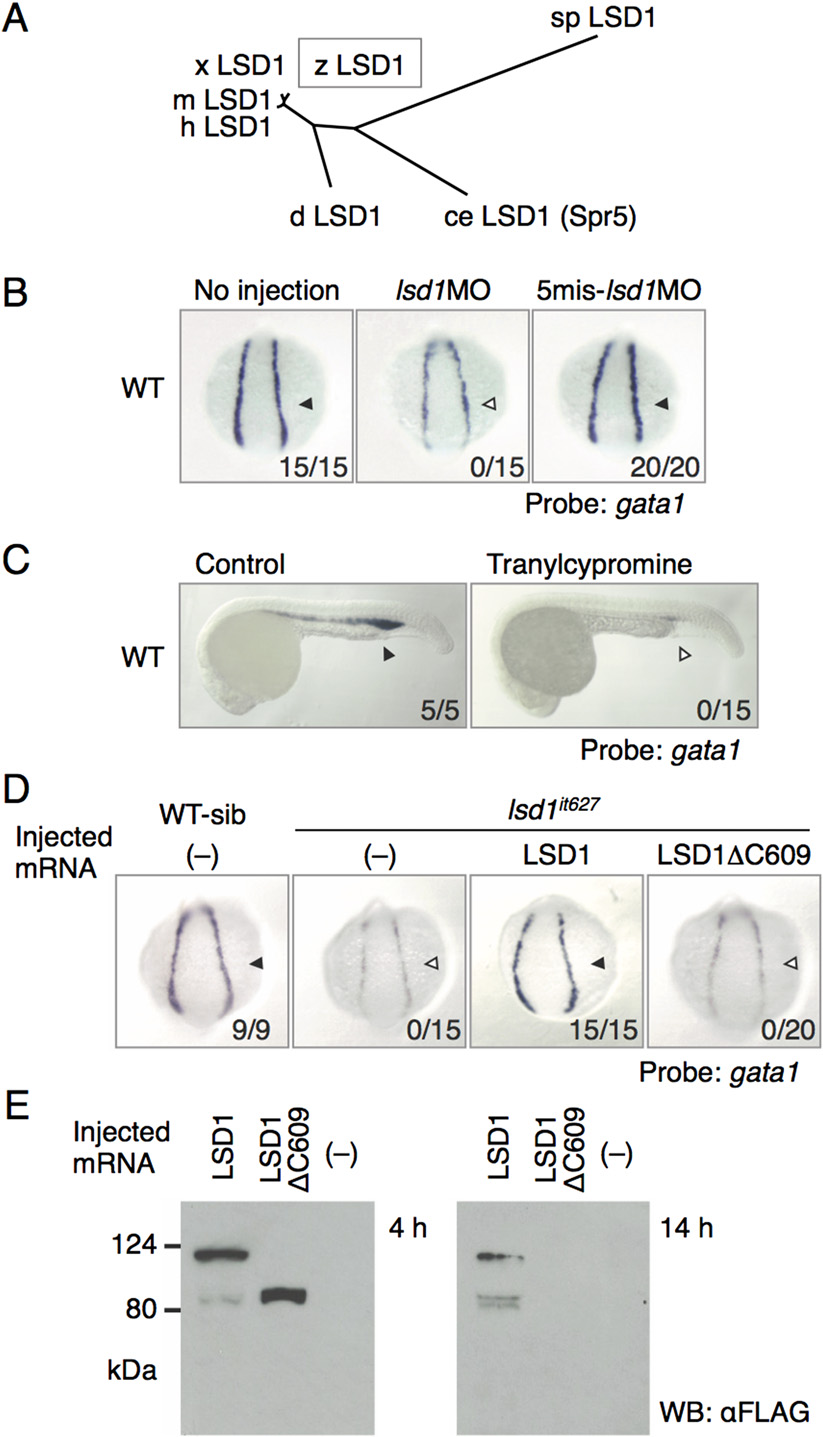
Fig. S1
Identification of LSD1 as a responsive gene for the it627 mutant. (A) Phylogenetic tree constructed by the neighbor-joining method for the full-length amino acid sequences of LSD1 proteins. ce, Caenorhabditis elegans; d, Drosophila melanogaster; h, human; m, mouse; sp, Schizosaccharomyces pombe; x, Xenopus laevis; z, zebrafish. (B) Expression of gata1 in the LPM at 14 hpf in WT embryos injected or not injected with 1 pmol of the indicated morpholino oligonucleotides. Arrowheads denote gata1 expression. Numbers indicate the percentage of embryos exhibiting strong gata1 expression. 5mis, 5-mismatch. (C) Effects of tranylcypromine on gata1 expression. The embryos were treated with 300 µM tranylcypromine in WT embryos from 12 hpf or earlier to 24 hpf. (D) Expression of gata1 at 14 hpf in WT or lsd1it627 embryos injected with or without mRNA encoding the indicated LSD1 proteins. WT-sib indicates WT or heterozygous mutants, and lsd1it627 designates homozygous mutants. (E) Expression and stability of overexpressed WT LSD1 and LSD1ΔC609 proteins in 4-hpf and 14-hpf embryos were examined by immunoblotting using anti-FLAG antibody. WB, Western blot.