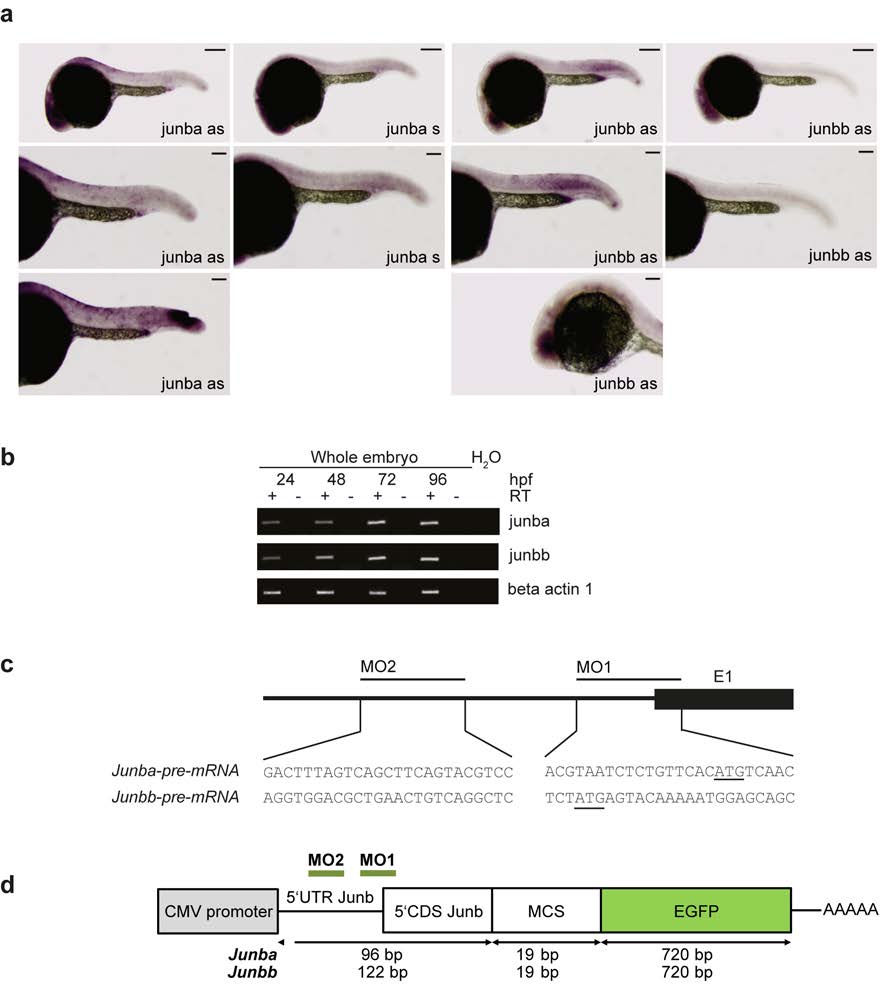
Fig. S1 Expression of junb in zebrafish embryos and junb morpholino design and its validation. (a) Whole-mount in situ hybridization of 24 hpf embryos shows expression of junba and junbb mRNA in the eye an danal placode. Junbb mRNA can also be detected in the mesoderm and vasculature. Staining with sense control probes is negative. Bottom panels show positive controls for detection of junb at sites of injury (left panel) and in the eye (right panel). Size bars correspond 250 µm in top panels, and 125 µm in middle and bottom panels. (b) Total RNA was isolated from developing zebrafish embryos at 24, 48, 72, and 96 hpf and used to determine transcript levels of junba and junbb by semi-quantitative PCR. Actb1 served as loading control. (c) Upper panel: junb-MOs were designed to block translation of junba and junbb mRNAs due to the lack of exon-intron boundaries. Lower panel: gene sequences of regions targeted by junba-MO1/MO2 and junbb- MO1/MO2 around or upstream of start of translation. Start codon is underlined. (d) Schematic depiction of the 0.1 kb-long junb:EGFP fusion constructs created to determine the knockdown efficiency of the junb-MOs. A 0.1 kb segment of the 5′UTR/5′CDS of zebrafish junba or junbb carrying the binding sites for both experimental morpholinos (green line: MO1, MO2) was amplified from genomic zebrafish DNA, subcloned into pGEM-T, and further cloned inframe into pEGFP-N1.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Sci. Rep.