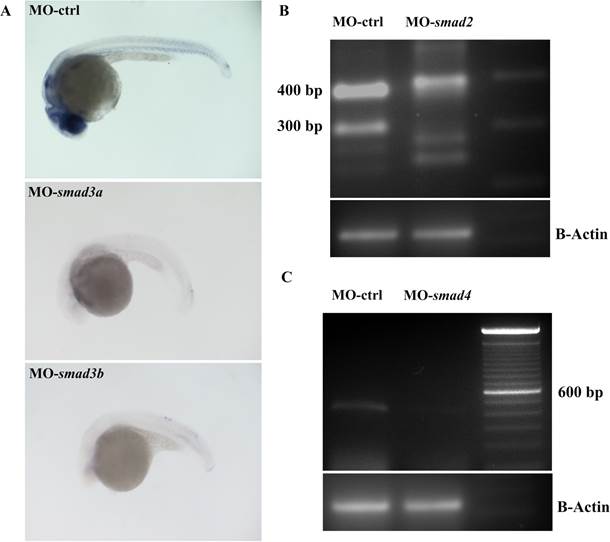
Fig. S3 smad2, 3a, 3b and 4 morpholinos are specific for their target. A, smad3a and 3b morpholinos are directed against the start codon of the corresponding target mRNAs. To validate them, control (MO-ctrl) smad3a (MO-smad3a) and smad3b (MO-smad3b) morphants were fixed and phospho-Smad3 production was revealed through an immunohistochemistry. phospho-Smad3 was drastically reduced in MO-smad3a and 3b. B–C, smad2 and 4 morpholinos act on a donor splicing site and their specificity was checked through a RT-PCR. cDNAs were normalized through β-Actin (B-Actin) amplification. B, RT-PCR amplification of RNA from control morphants (MO-ctrl) and smad2 morphants (MO-smad2). There are two splicing variants for smad2 named NSDART00000147653 and NSDART00000044756. smad2 cDNA was amplified with a forward primer in exon 1 and a reverse primer in exon 3. MO-ctrl gave the expected bands of 383 bp (NSDART00000147653) and 304 bp (NSDART00000044756), while in MO-smad2 these PCR products disappeared. In smad2 morphants additional bands clearly appeared caused by the activation of a cryptic splicing site. C, RT-PCR amplification of RNA from control morphant (MO-ctrl) and smad4 morphant (MO-smad4). smad4 cDNA was amplified with a forward primer in exon 3 and a reverse primer in exon 5. In MO-ctrl we observed the expected single band of 438 bp, while in MO-smad4 this PCR product was almost completely absent.
Reprinted from Developmental Biology, 396(1), Casari, A., Schiavone, M., Facchinello, N., Vettori, A., Meyer, D., Tiso, N., Moro, E., Argenton, F., A Smad3 transgenic reporter reveals TGF-beta control of zebrafish spinal cord development, 81-93, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.