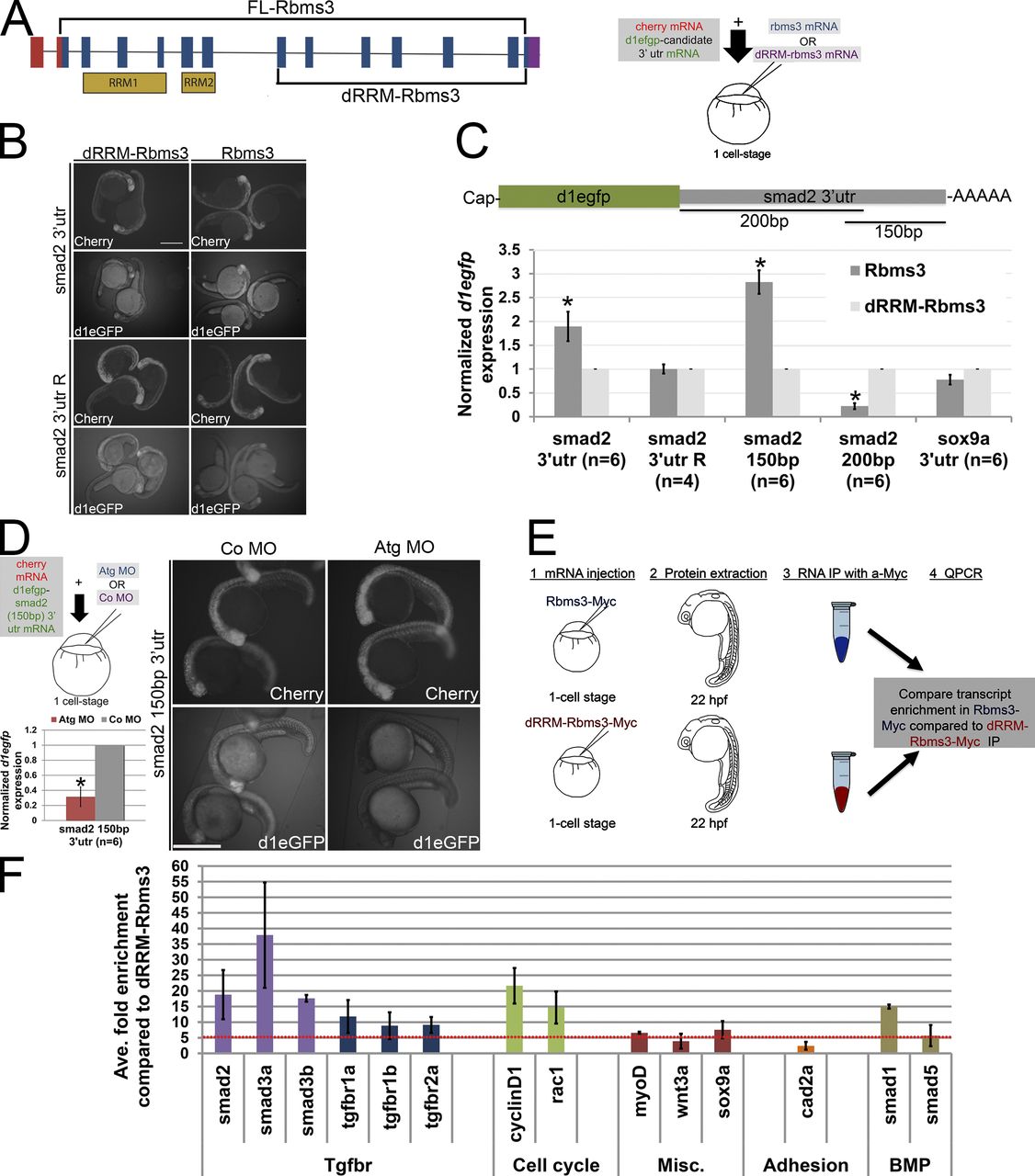
Fig. 8 Smad2 transcripts are a target of Rbms3. (A) dRRM-Rbms3 lacks the putative RNA-binding domains (RRM; lower bracket, left). A schematic showing an in vivo stability assay is shown on the right. (B and C) Fluorescent images (B) and QPCR (C; d1egfp normalized to cherry) showing that full-length Rbms3 treatment stabilizes d1egfp-smad2 32 utr and d1egfp-smad2 (150bp) 32 utr reporters. (D) Schematic shows the experiment (top left), QPCR (d1egfp normalized to cherry; bottom left), and fluorescent images (right) showing that Atg MO treatment destabilizes d1egfp-smad2 (150bp) 32 utr. (B and D) Images were taken under same brightness/contrast conditions. (E) Schematic of RIP experiment. (F) QPCR showing transcripts enriched (more than fivefold, red dotted line) for full-length Rbms3 (n = 2–3 experiments). Error bars indicate SEM (C) and SD (D and F). n = RNA from individual embryos (C and D). *, P < 0.05 (Student’s t test). Bars, 500 µM.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Creative Commons License (Attribution–Noncommercial–Share Alike 3.0 Unported license, as described at http://creativecommons.org/licenses/by-nc-sa/3.0/ ).
Full text @ J. Cell Biol.