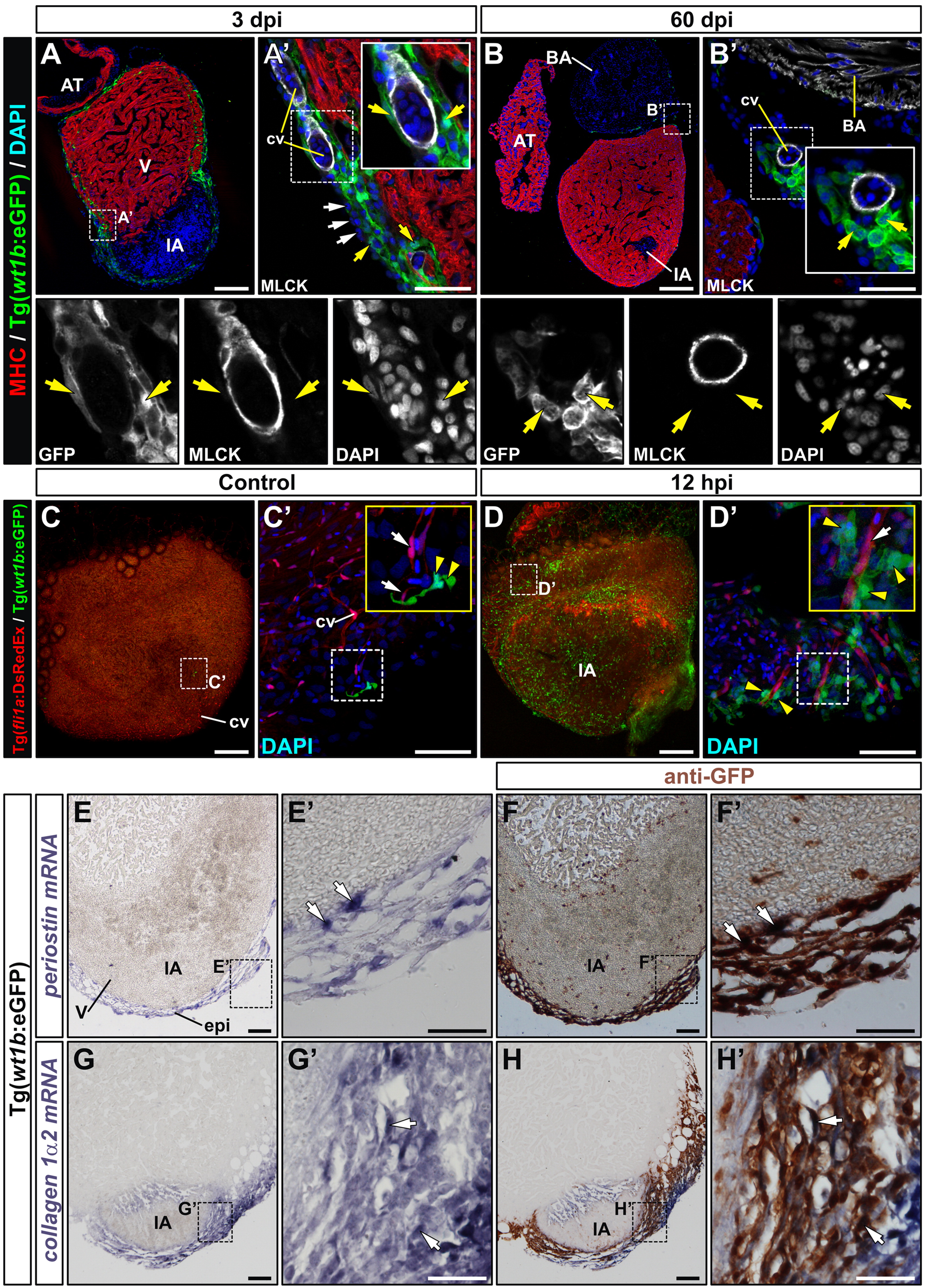
Fig. 2 wt1b:eGFP cells give rise to perivascular cells and express fibroblast marker genes upon cryoinjury. (A) and (B) Immunohistochemistry on sections of cryoinjured Tg(wt1b:eGFP) hearts, using antibodies against myosin heavy chain (red) to detect myocardium and GFP (green) to detect wt1b:eGFP+ cells. A′-B′ are zoomed images of boxed areas in A and B, additionally showing myosin light chain kinase (MLCK, white) to detect smooth muscle cells surrounding coronary endothelial cells. The bottom row shows three separate channels for the zoomed regions. (A′) At 3 dpi, GFP+ cells surround but do not co-localize with MLCK+ cells. Note that not all epicardial cells are GFP+ (white arrows). (B′) At 60 dpi the few GFP+ cells visible in the cryoinjured heart are again associated with coronary vessels. Yellow arrows mark GFP+ cells. (C) and (D) Whole mount fluorescence imaging in control (C-C′) and 12 hpi cryoinjured hearts (D,D′) from the Tg(wt1b:eGFP)/Tg(fli1a:DsRedEx) line. Zoomed views show DAPI nuclear counterstaining. Endothelial cells are visible by DsRed fluorescence. White arrows mark endothelial cells and yellow arrowheads highlight wt1b:GFP+ cells in close apposition. (E-H) Sections of cryoinjured Tg(wt1b:eGFP) hearts (3 dpi) hybridized with riboprobes for periostin and collagen 1 alpha 2 mRNAs. In panels F and H, sections were immunostained for GFP after ISH. Arrows mark GFP+ cells co-labelled with the ISH probe. Abbreviations are as in previous figures. A′-H′ are zoomed views of the boxed areas in E-H. Bars, 200 μm (full views), 50 μm (magnifications).
Reprinted from Developmental Biology, 370(2), Manuel González-Rosa, J., Peralta, M., and Mercader, N., Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration, 173-186, Copyright (2012) with permission from Elsevier. Full text @ Dev. Biol.