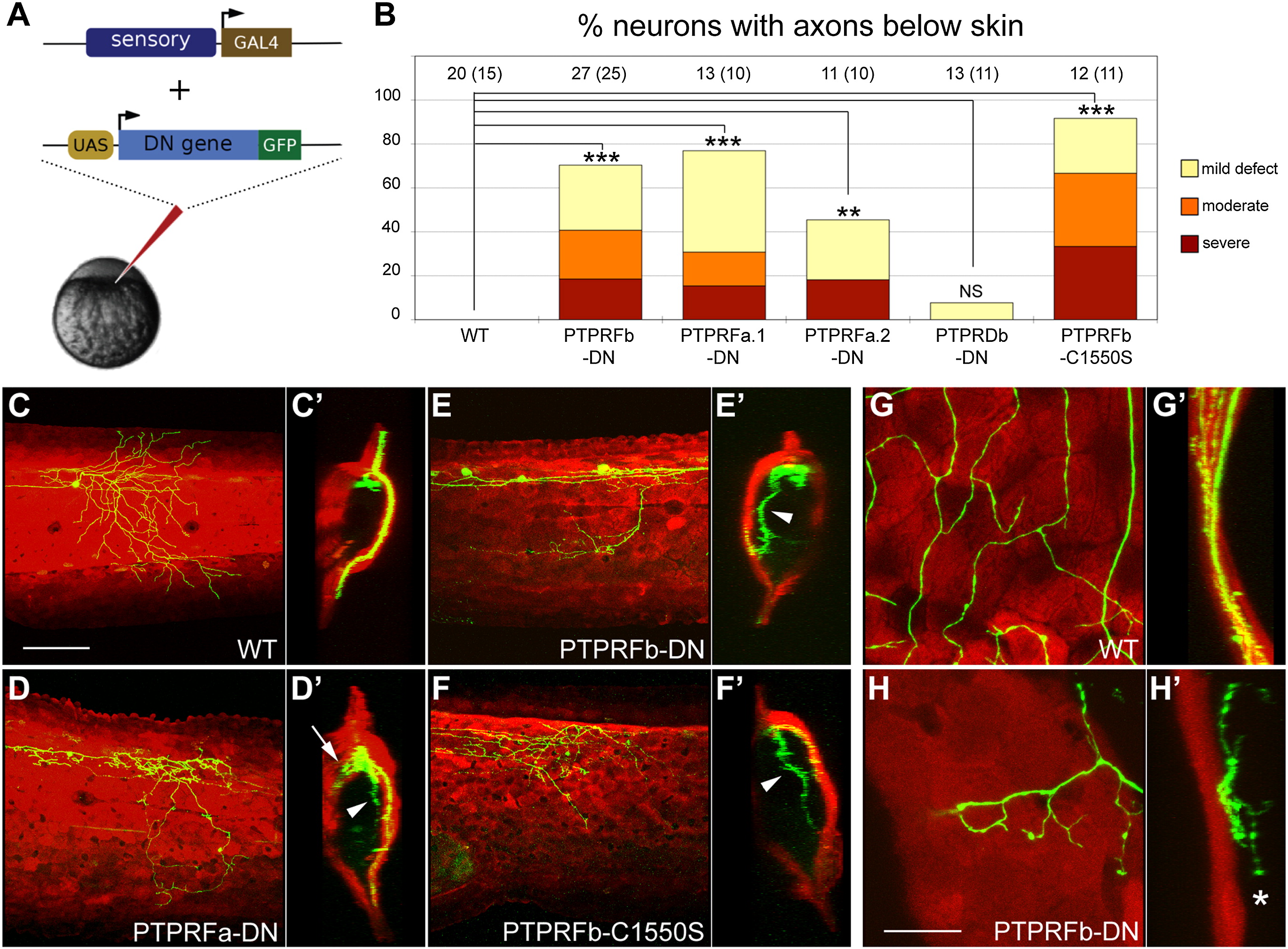
Fig. 3
Rohon-Beard Neurons Expressing Dominant-Negative LAR Family Proteins Failed to Innervate Skin (A) A sensory neuron-specific promoter was used to drive the GAL4-VP16 activator [37]. Putative dominant-negative (DN) versions of each gene, created by replacing their cytoplasmic domains with GFP, were expressed under the control of the Gal4 upstream activation sequence (UAS). Coinjection of these transgenes at the one-cell stage caused transient mosaic expression in somatosensory neurons. (B) Frequency of RB neurons expressing indicated transgenes with peripheral axons below the skin. Number of neurons are listed at the top of each bar; number of fish in parentheses. Data were analyzed with two-sided Fisher′s exact test and computed with R. ***p < 0.001; **p < 0.01; NS, not significant. Defects were scored as “mild” if one or a few axon branches were below the skin, “moderate” if many axon branches were below the skin, and “severe” if the entire peripheral arbor was below the skin. (C–F) Confocal projections of lateral views of RB neurons expressing GFP only (WT) (C), PTPRFa.1-DN (D), PTPRFb-DN (E), and PTPRFb-C1550S (F). Ninety degree rotations of the images in (C)–(F) provide cross-section views of embryos (C2–F2), respectively. krt4:dsRed was used to visualize the skin. Arrowheads indicate axons innervating internal tissues; arrow indicates axons innervating the contralateral side of the body. Scale bar represents 100 μm. (G–H) Higher magnification of lateral view RB neurons expressing GFP only (WT) (G), and PTPRFb-DN (H). Ninety degree rotations are shown in (G2–H2). Star indicates axon arborizing just beneath the skin. Scale bar represents 20 μm. See also Figure S3.