Fig. 3
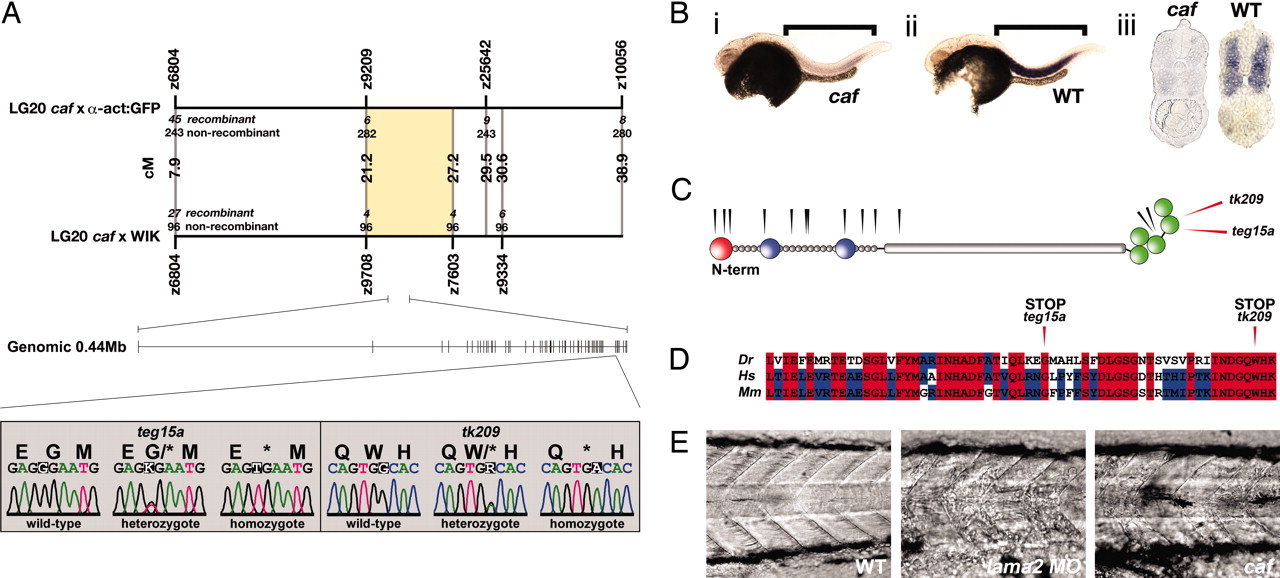
The caf phenotype is a result of mutations in the zebrafish homologue of the LAMA2 gene. (A) Initial bulked segregant analysis implicated markers z6804 and z10056 as being linked to the caf phenotype. Further fine mapping resolved flanking markers z9708 and z7603, delineating a candidate genomic region of H0.89 Mb, containing sequence with homology to human LAMA2. Single base changes in both cafteg15a and caftk209 were found in the homologue of human exon 60, resulting in the introduction of premature stop codons to the zebrafish lama2 ORF. (B) In situ hybridization to the lama2 mRNA at 48 hpf shows expression in the myotomal muscle. Strikingly, there is a severe reduction of message in caf homozygous embryos, suggestive of nonsense-mediated decay. Brackets indicate region of myotome. (i) Homozygous caf embryo, (ii) wild-type embryo, and (iii) transverse methacrylate sections through the trunk of caf (Left) and wild-type (Right) embryos. (C) Structure of the human LAMA2 protein. The positions of mutations that result in the MDC1A disease are indicated by black arrowheads. The homologous positions of the cafteg15a and caftk209 stop codons are indicated by red arrowheads. Both caf mutations occur in close proximity to known human MDC1A mutations, within the globular domain, which is essential for dystroglycan binding. Domains represented are laminin VI (red), laminin IV (blue), EGF-like (gray), and globular (green). (D) Amino acid alignment between zebrafish (Dr), human (Hs), and mouse (Mm) sequences for the mutant exon. Both cafteg15a and caftk209 mutations occur within completely conserved amino acids. (E) Injection of antisense morpholino oligonucleotides against the lama2 mRNA phenocopies the caf homozygous mutants. Seventy-two hpf, lateral view, anterior to the left.