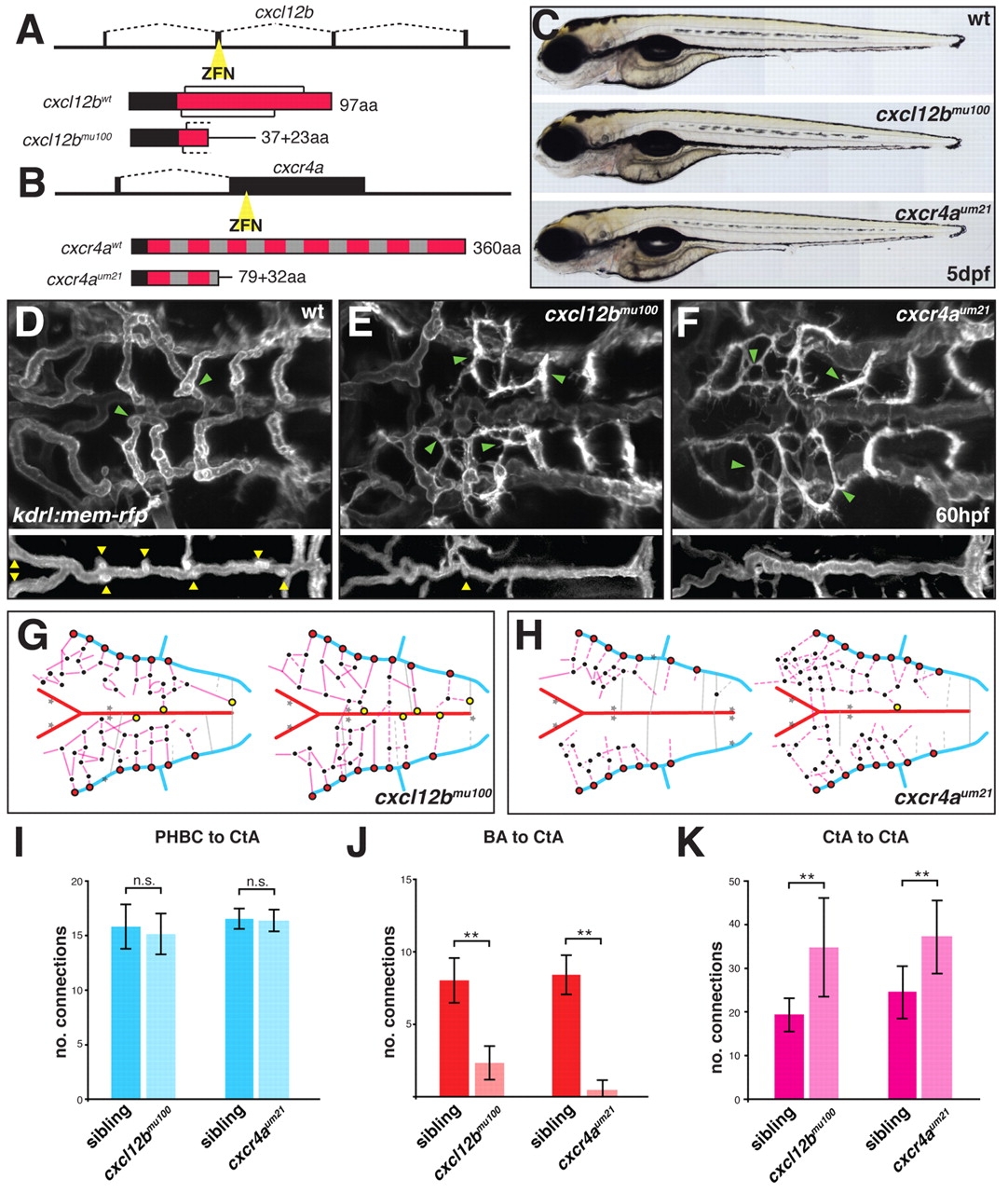
Fig. 4
cxcl12b and cxcr4a are required for hindbrain vascular patterning. (A,B) Schematic representation of cxcl12b (A) and cxcr4a (B) mutant generation using zinc-finger nucleases (ZFNs). Black boxes in the gene structure represent exons and dashed lines represent introns. Yellow triangle indicates the position of the ZFN targeting site. Protein structures are displayed as red boxes. Black brackets indicate the position of conserved cysteine bridges for Cxcl12b. Black boxes in the protein structure represent the signal peptide for secretion (Cxcl12b) or membrane targeting (Cxcr4a). Gray boxes indicate the position of transmembrane helices for Cxcr4a. aa, amino acid. (C) Brightfield images of wt, cxcl12bmu100 and cxcr4aum21 homozygous mutant zebrafish at 5 dpf. (D-F) Maximal intensity projections of confocal z-stacks of kdrl:mem-rfp transgenic embryos at 60 hpf. wt (D), cxcl12bmu100 (E) and cxcr4aum21 (F) homozygous mutants are shown. Yellow arrowheads indicate CtA to BA connections; green arrowheads indicate CtA to CtA connections. (G,H) Schematic representation of the hindbrain vascular network in two individual cxcl12b mu100 (G) and cxcr4aum21 (H) homozygous mutant embryos based on confocal z-stacks of kdrl:gfp transgenic embryos. Color coding as in Fig. 1C. Gray stars indicate missing conserved vascular connections. (I-K) Quantitative analysis of hindbrain patterning in wt, cxcl12bmu100 and cxcr4aum21 homozygous mutant zebrafish. Vessel connections were counted from confocal z-stacks of kdrl:gfp transgenic embryos. **, P<0.05; n.s., not significant (Mann-Whitney U test). n=6 embryos per group. Error bars represent s.d.