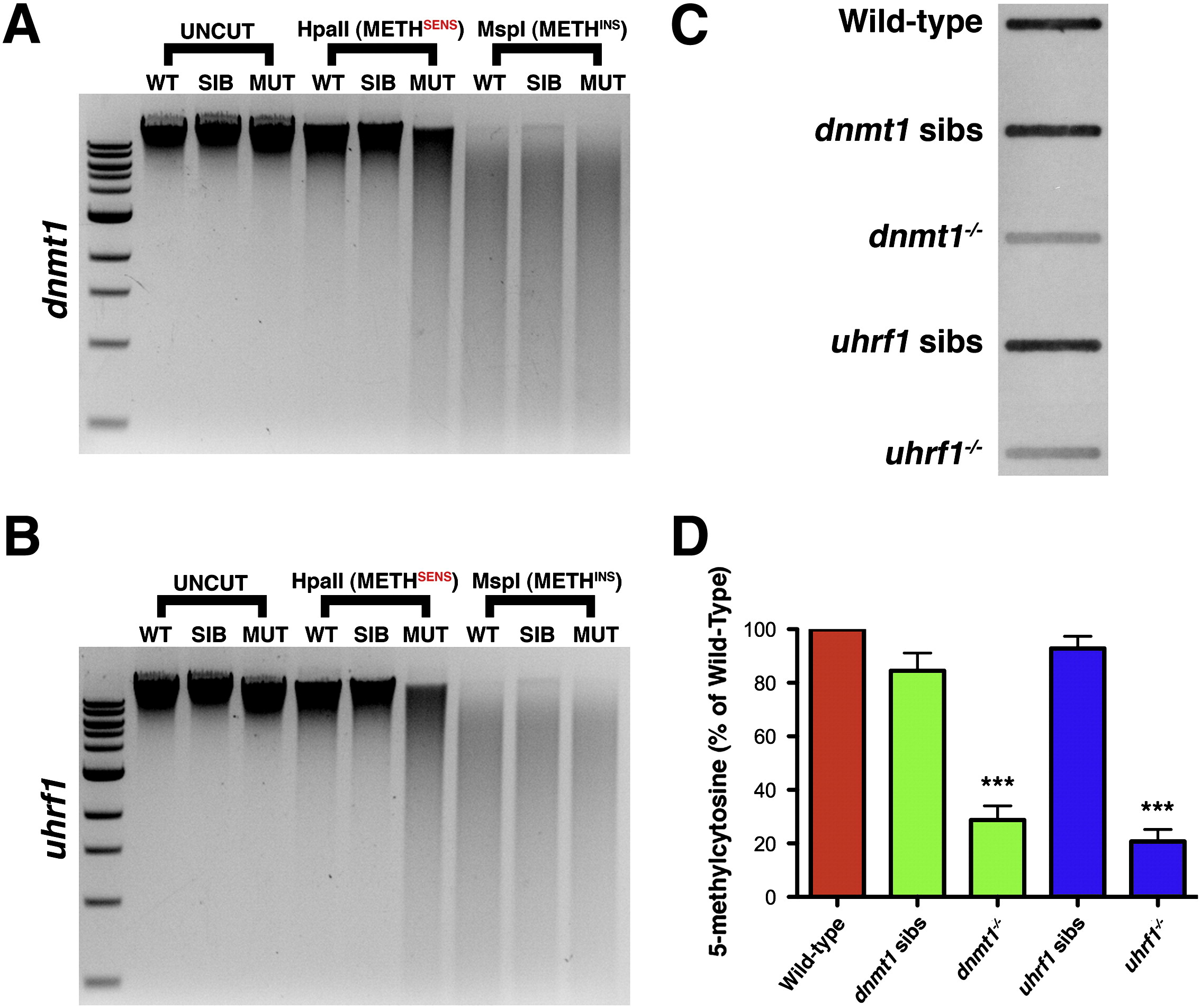
Fig. 1
uhrf1 mutants possess hypomethylated genomic DNA. Genomic DNA methylation assay in which 750 ng of genomic DNA is digested by a methylation-sensitive enzyme (HpaII), a methylation-insensitive enzyme (MspI), or a mock digestion with no enzyme present. (A) Genomic DNA isolated from Wild-type AB, dnmt1 siblings or dnmt1 mutant embryos demonstrates the efficacy of the assay. Genomic DNA is digested by the methylation-sensitive HpaII to a greater extent in the mutant embryos than in siblings or wild-types. (B) Genomic DNA isolated from Wild-type AB, uhrf1 siblings or uhrf1 mutant embryos; uhrf1 mutant DNA is digested by the methylation sensitive HpaII to a greater extent than sibling or wild-type DNA. (C) SouthWestern assay to quantify 5-methylcytosine levels on genomic DNA. 2 µg of genomic DNA is extracted from the indicated group of embryos, loaded onto a membrane and probed with anti-5-methylcytosine antibody. (D) Quantification of 5-methylcytosine levels (n = 8 trials; *** p < 0.00002). Error bars represent s.e.m.
Reprinted from Developmental Biology, 350(1), Tittle, R.K., Sze, R., Ng, A., Nuckels, R.J., Swartz, M.E., Anderson, R.M., Bosch, J., Stainier, D.Y., Eberhart, J.K., and Gross, J.M., Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens, 50-63, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.