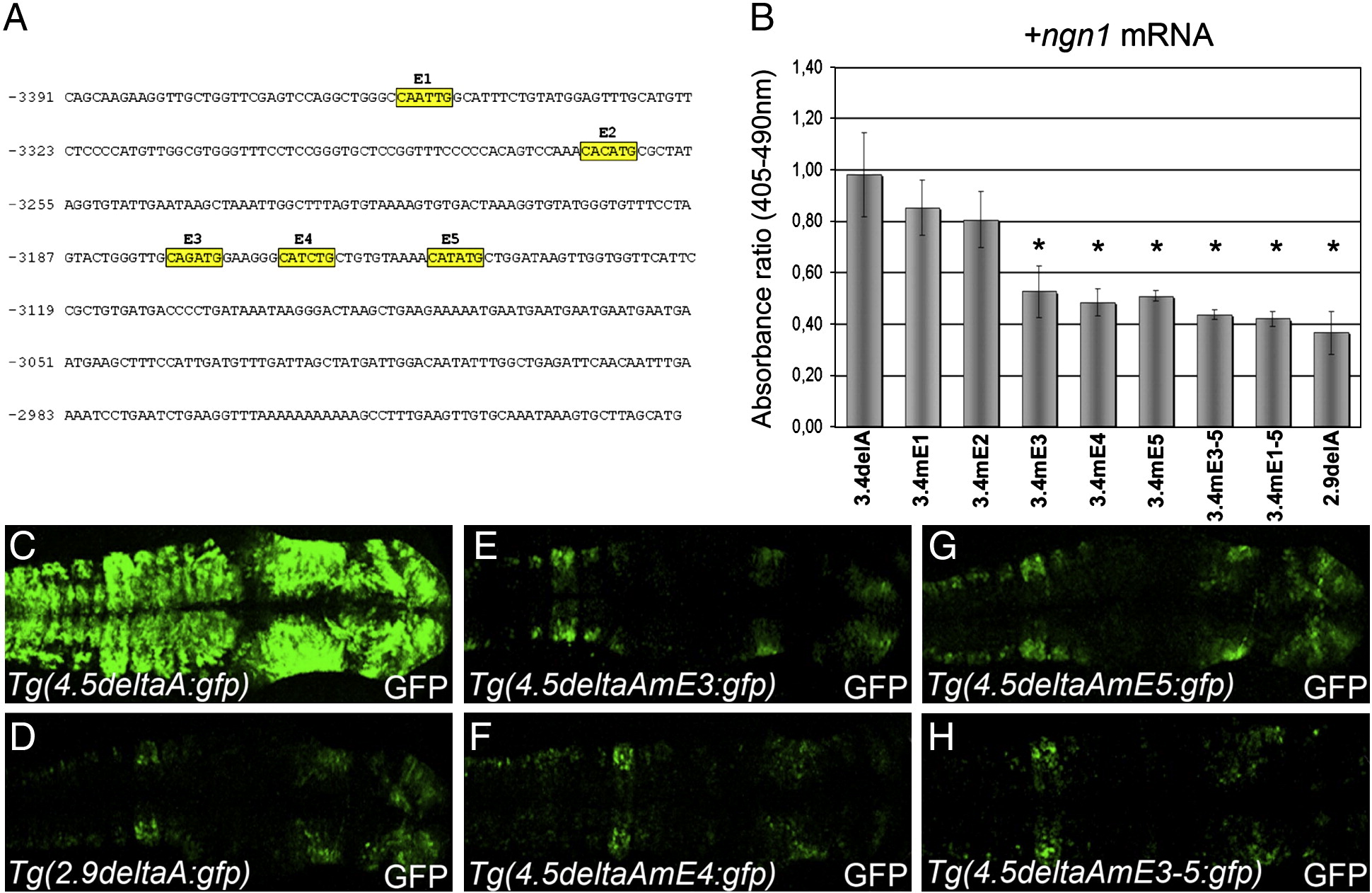
Fig. 5 E-boxes are required for the transcriptional regulation of deltaA by Ngn1.
(A) Nucleotide sequence of genomic DNA located between -3.4 and -2.9 kb upstream of the DeltaA coding region. Yellow boxes highlight the five E-boxes in this sequence. (B) Histogram showing the ratio of CAT levels after the mis-expression of various promoter:reporter constructs and synthetic ngn1 mRNA. CAT expression levels were normalized against LacZ driven off the 7.1deltaA:LacZ fragment. (C–H) Confocal projections after immunohistochemical detection of GFP showing expression in the anterior neural tube in a series of stable transgenic embryos at 24 hpf. Whereas Tg(4.5deltaA:GFP) embryos show strong and widespread GFP expression (C) the Tg(2.9deltaA:GFP) transgene drives weaker GFP expression in a more restricted pattern (D). Transgenic lines where individual E-box (E–G) or the three clustered E-boxes were mutated (H) display GFP expression similar to Tg(2.9deltaA:GFP). Embryos are viewed dorsally and mounted with anterior to the right. Error bars represent s.d. *P < 0.05; **P < 0.001; ***P < 0.0005 using a t-test.
Reprinted from Developmental Biology, 350(1), Madelaine, R., and Blader, P., A cluster of non-redundant Ngn1 binding sites is required for regulation of deltaA expression in zebrafish, 198-207, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.