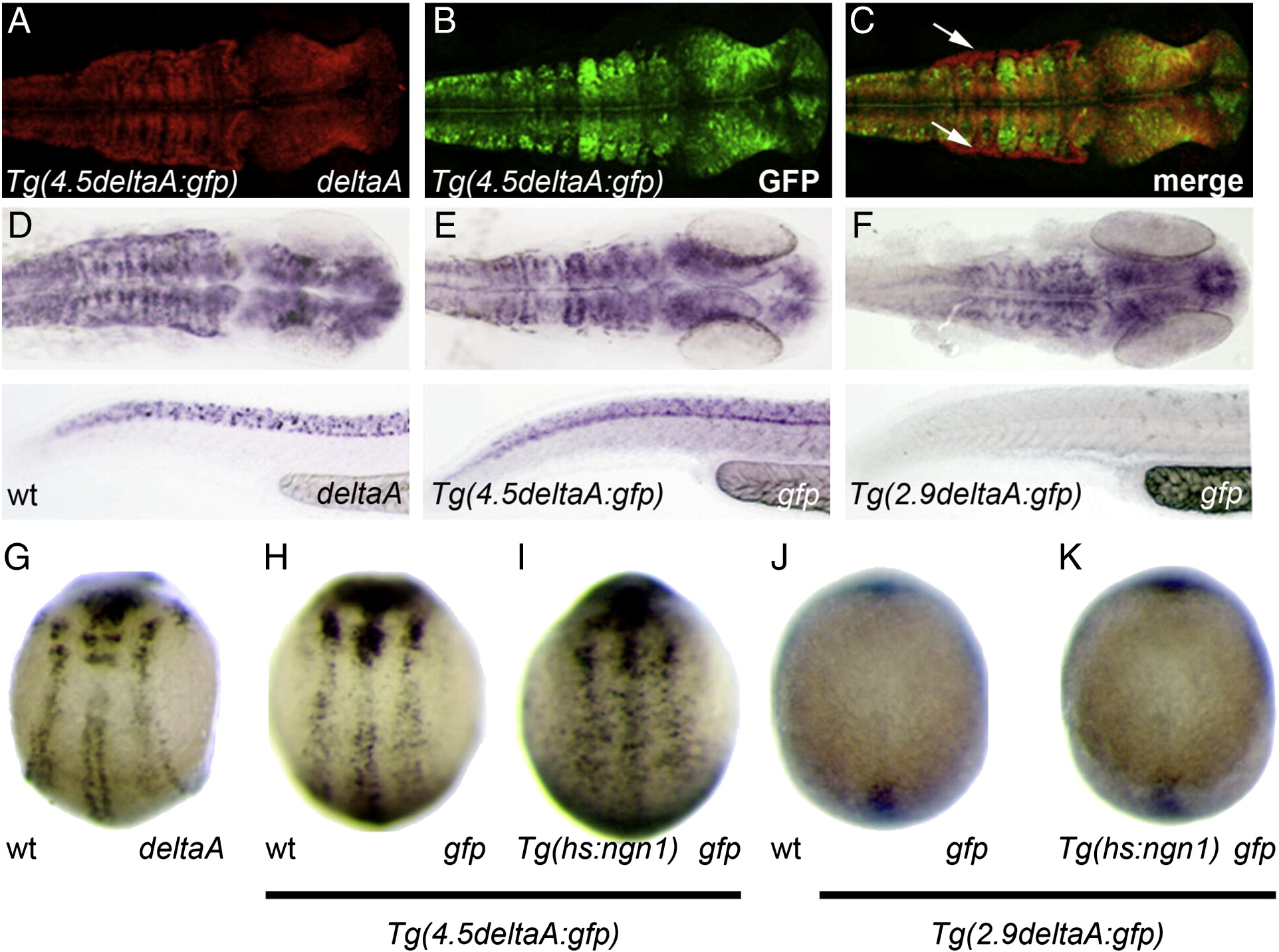
Fig. 4 A ngn-regulated cis-regulatory module is located between -2.9 and -4.5 kb upstream of deltaA.
(A–C) Confocal projections of double in situ/immunolabelling showing extensive overlap in the expression of endogenous deltaA mRNA and GFP protein expression in Tg(4.5delA:GFP) embryos at 24 hpf; a notable exception is the dorsal limit of the hindbrain that expresses deltaA but where GFP is not detected (white arrows in C). (D–F) Whole-mount in situ hybridization against deltaA (D) or gfp in Tg(4.5delA:GFP) (E) versus Tg(2.9delA:GFP) (F) transgenic embryos at 24 hpf. The shorter transgene drives gfp expression in a significantly more restricted pattern. (G, H) The expression of endogenous deltaA and GFP expression in Tg(4.5delA:GFP) embryos at 10 hpf are highly similar. (H–K) Mis-expression of Ngn1 induces ectopic expression of gfp in 10 hpf Tg(4.5deltaA:GFP) embryos but not in stage-matched Tg(2.9deltaA:GFP) embryos. Embryos at 24 hpf are mounted with anterior to the right; the brain is viewed dorsally and the trunk and tail are viewed laterally with dorsal part up; at 10 hpf, embryos are viewed dorsally with anterior up.
Reprinted from Developmental Biology, 350(1), Madelaine, R., and Blader, P., A cluster of non-redundant Ngn1 binding sites is required for regulation of deltaA expression in zebrafish, 198-207, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.