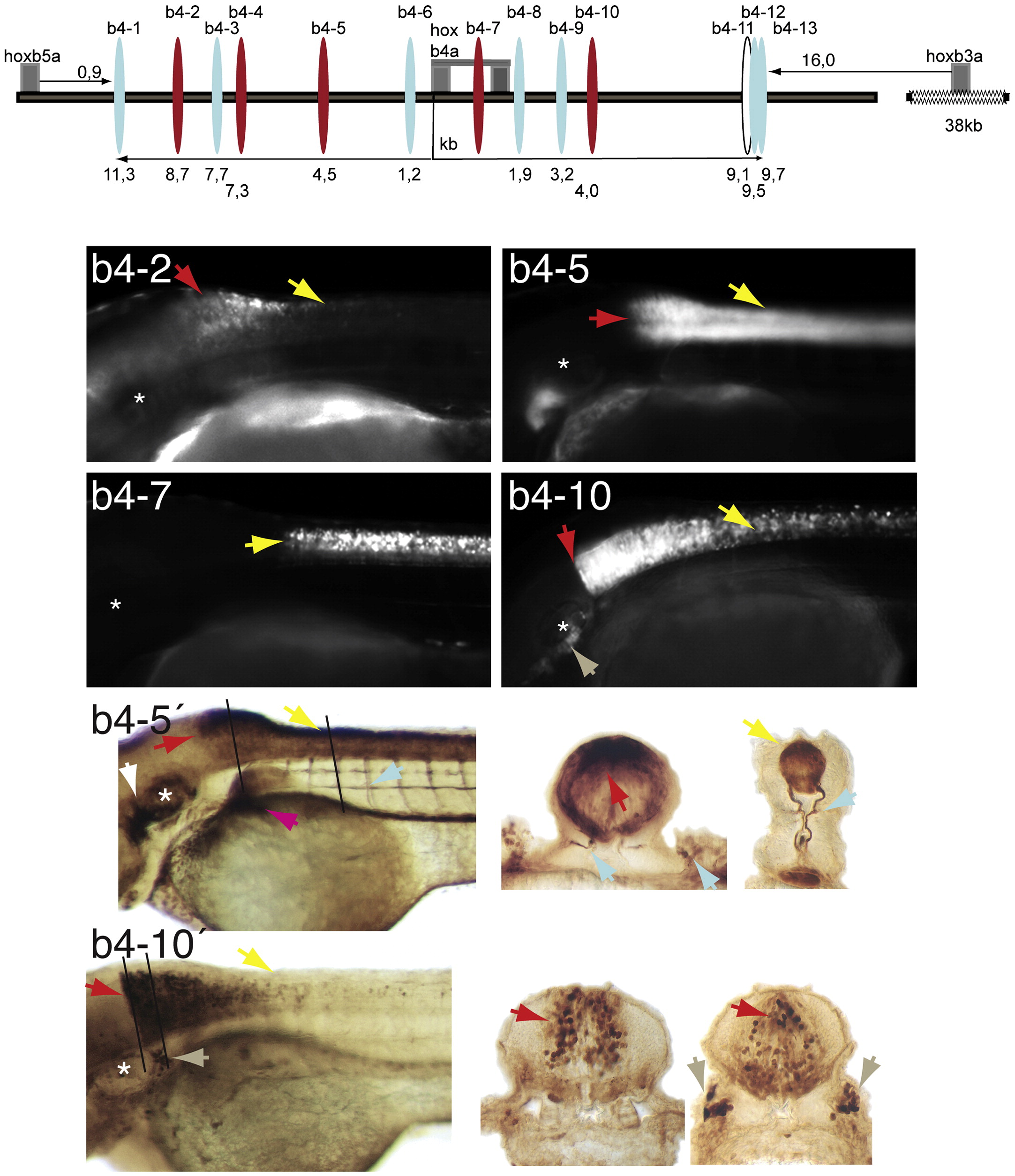
Fig. 5 Schematic of the hoxb5a to hoxb3a genomic region. Tested elements are indicated as ovals (red: tissue-specific enhancer activity, blue: unspecific, white: no activity) with its distance from the exons in kb. The regulatory activity of the elements is illustrated by representative larvae at 2 to 4 dpf. The asterisk marks the level of the inner ear. The arrows follow a color code: red, hindbrain; yellow, spinal cord; brown, cranial ganglia; light blue, neuronal projection; white, migrating neural crest. b4-2. GFP expression in the posterior hindbrain, extending with decreasing intensity into the anterior spinal cord. b4-5. Larva with strong and broad GFP expression in the hindbrain r7 and r8 extending posterior into the spinal cord. GFP is also visible in a pectoral fin neuron, in the developing pharyngeal arches and a cranial ganglion. b4-5′. Anti-GFP stained larva with sections at the level of the 2nd somite and the spinal cord. The level of sections are indicated by the black lines. Strong GFP expression is indicated in the dorsal and lateral domains of hindbrain r7 and r8 with projections into the pectoral fins. In the spinal cord the signal is also stronger dorsally with two clusters of motoneurons projecting laterally along the adjacent somites. b4-7. Larva with strong GFP expression in the entire spinal cord. b4-10. Larva with GFP expression in the hindbrain r7 and r8 with a sharp anterior expression boundary at r6. Expression extends posterior into the spinal cord. GFP is also visible in cranial ganglia around the inner ear. b4-10′. Anti-GFP stained larva of the same transgenic line with sections through the hindbrain r6/7 and r7, which shows the distribution of the GFP positive cells in the medial hindbrain and in cells of the posterior lateral line ganglion.
Reprinted from Developmental Biology, 340(2), Punnamoottil, B., Herrmann, C., Pascual Anaya, J., D'Aniello, S., Garcia-Fernàndez, J., Akalin, A., Becker, T.S., and Rinkwitz, S., Cis-regulatory characterization of sequence conservation surrounding the Hox4 genes, 269-282, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.