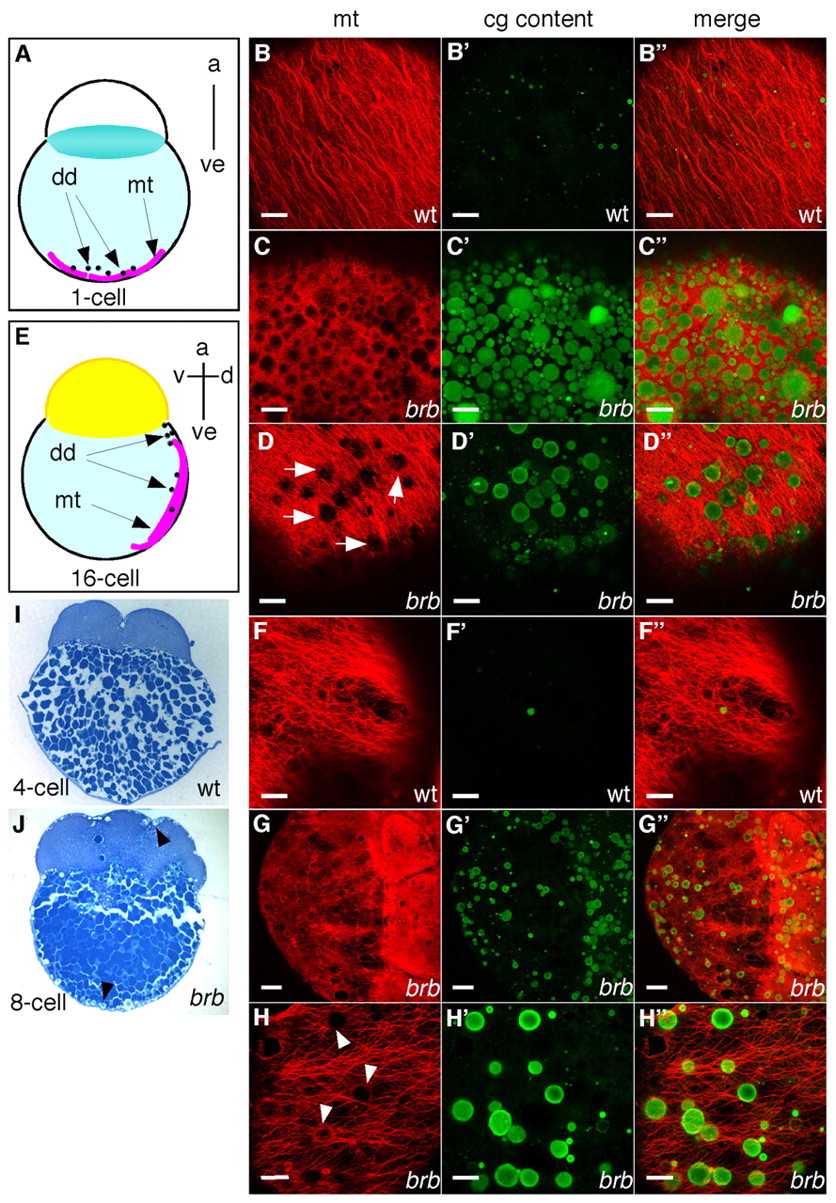
Fig. 6 brom bones mutants display cortical microtubule defects associated with impaired CGE. (A,E) Schematic diagrams showing parallel arrays of microtubules in the vegetal cortex at the 1-cell stage (A) and in the lateral yolk cytoplasmic layer at the 16-cell stage (E). (B-D″,F-H″) Zebrafish embryos were double-stained to show microtubules (red) and CG contents (green). Confocal image stacks of 6 μm (B-F″,H-H″) and 80 μm (G-G″). (B-D″) Embryos at 20 mpf; vegetal pole views. (B-B″) Microtubules and traces of CG content in wild-type embryos (n=22). Microtubules and persisting CGs in severely affected (C-C″, 16%, n=88) and intermediately affected (D-D″, 57%) brom bones mutant embryos. The remaining mutant embryos showed similar microtubule arrays to wild type (27%, data not shown). (F-H″) Embryos at the 16-cell-stage; lateral views. (F-F″) Cortical microtubules in wild-type embryos (n=35). (G-G″) In brom bones mutants the microtubule network was disrupted by circular profiles, which correspond to non-exocytosed CGs (71%, n=80 mutant embryos). The remaining mutant embryos showed a normal microtubule network (29%, data not shown). (H-H″) Observation at high magnification shows that the circular profiles (arrowheads) interrupting the long microtubule arrays in mutant embryos are CGs. (I,J) CGs were not observed in wild-type embryos during early cleavage stages (I), but were found in mutant embryos beneath the plasma membrane (J, arrowheads). Sagittal histological sections (5 μm) were stained with Toluidine Blue to show CGs. a, animal pole; ve, vegetal pole; d, dorsal; v, ventral; dd, dorsal determinant; mt, microtubules. Scale bars: 20 μm in B-F″,H-H″; 50 μm in G-G″.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development