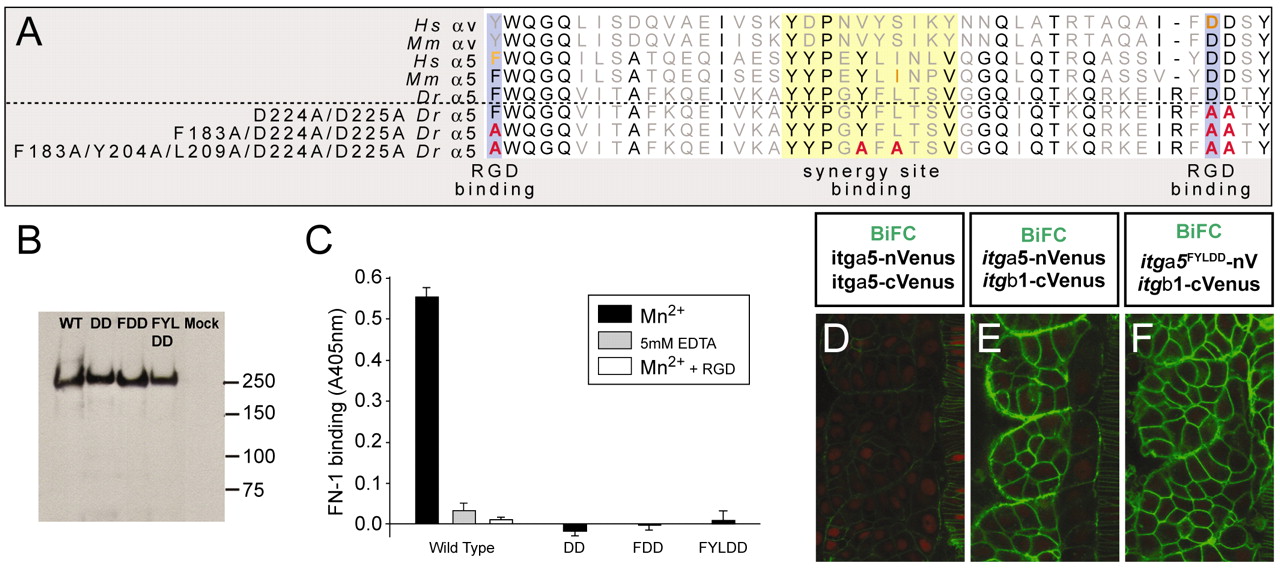
Fig. 2 Ligand-binding-deficient variants of Itgα5. (A) An alignment of a portion of the β-propeller domain of human (Hs), mouse (Mm) and zebrafish (Dr) Integrin α5 and αV. Residues known to be necessary for RGD binding (blue) and synergy site binding (yellow) are indicated. The orange residues have been analyzed experimentally in the corresponding Integrin. The sequences of the modified zebrafish Itgα5 are shown with the altered residues in red. (B) Western blotting of wild-type (WT) and mutant (DD, FDD and FYLDD) recombinant-soluble α5β1-Fc Integrins, showing that in each case a heterodimer is formed of ∼240 kDa. This band is not observed in proteins purified from the supernatants of mock-transfected cells. (C) Ligand-binding assay. Binding of a recombinant fragment of zebrafish Fn1 to wild-type or mutant (DD, FDD and FYLDD) α5β1-1-Fc was measured in a solid-phase assay in the presence of 1 mM Mn2+ to activate the Integrin (black bars). To demonstrate specificity of Fn1 binding to the wild-type Integrin, the interaction was also measured in the presence of cyclic RGD peptide (white bar) or 5 mM EDTA (gray bar). (D-F) BiFC demonstrates that the mutant Itgα5FYLDD forms a heterodimer with Itgβ1 on the cell cortex in vivo. Negative (D) and positive (E) controls are also shown. Anterior is up and medial is to the right.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development