Fig. 6
Motif-based search for functional zebrafish orthologs and SoxE positive enhancers.
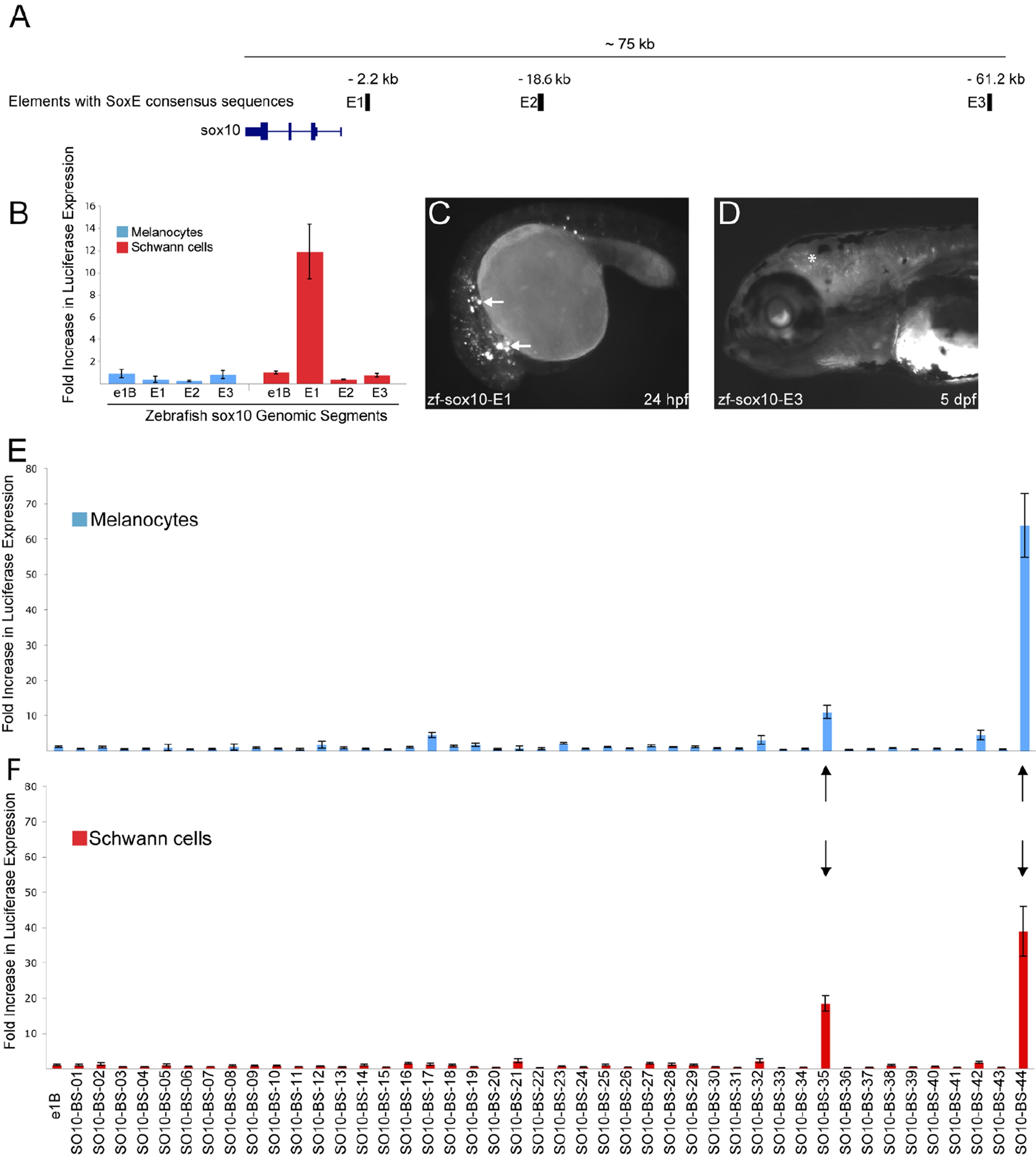
A) A computational search for dimeric SoxE consensus sequences in a 100 kb window upstream of the zebrafish sox10 gene resulted in three hits (pale blue bars). The distance from the sox10 TSS is denoted above each enhancer (E1–E3). B) A 500 bp region surrounding each dimeric SoxE consensus sequence was tested for in vitro enhancer activity in a melanocyte cell line (blue bars) and a Schwann cell line (red bars). Each fragment is compared against a control (pE1b with no insert) and error bars indicate the standard deviation. C) zf-sox10-E1 directs reporter expression in scattered cells throughout the 24 hpf, G0 embryo, consistent with migrating neural crest cells (white arrows). D) zf-sox10-E3 directs reporter expression in cranial oligodendrocytes (asterisks) at 5 dpf in G0 embryos. E and F) Data mining of the mouse genome for highly conserved dimeric SoxE consensus sequences in the intron of genes expressed in melanocytes identified 44 genomic segments. In vivo enhancer activity was tested in cultured melanocytes (E) and Schwann cells (F). Only two segments drove luciferase expression above a 10-fold threshold in both cell lines (black arrows). Error bars, SD.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ PLoS Genet.