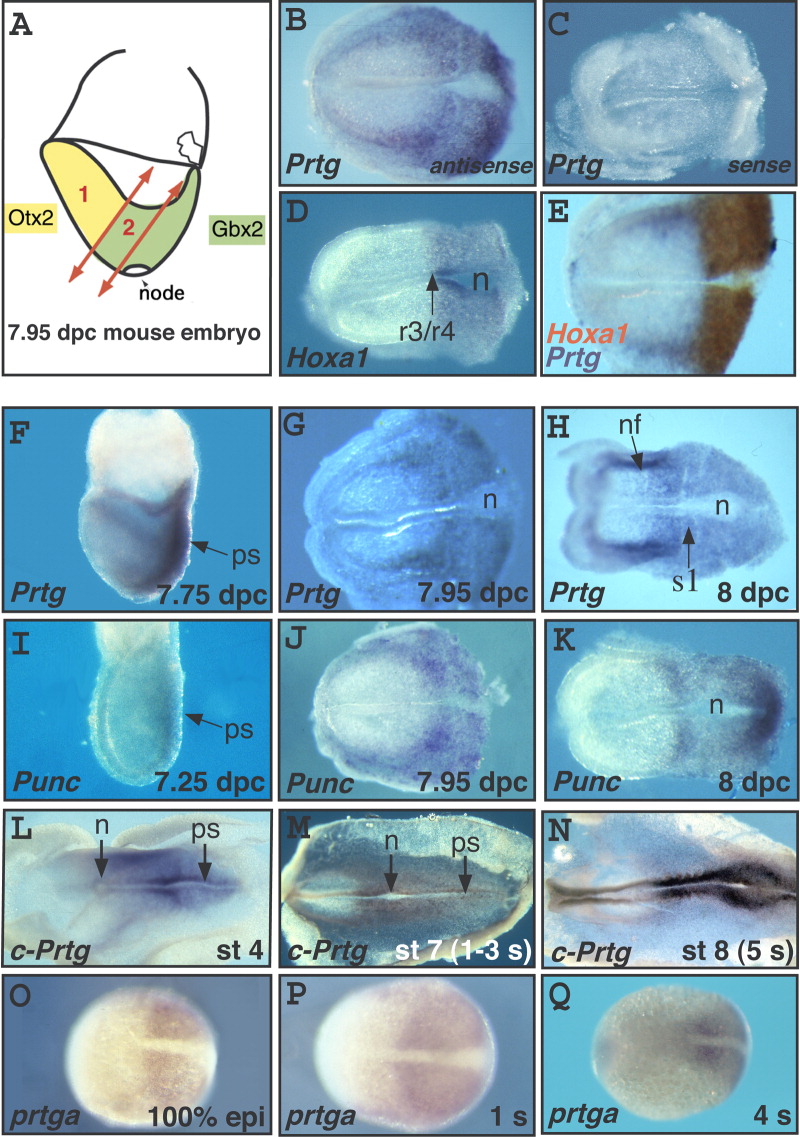
Fig. 1 Comparative analysis of Prtg expression at the end of gastrulation. A: Schematic representation of a mouse embryo at the headfold (7.95 days postcoitum [dpc]) stage, showing the two regions (1 and 2) from which RNA was extracted to build the subtracted libraries. B-D: In situ hybridization (ISH) on mouse embryos at the headfold stage with probes corresponding to the most frequently found genes from the posterior library: Prtg, a novel gene, represented by clone P1-50 (B and C for antisense and sense probe, respectively) and Hoxa1 (D). E: Double ISH with Prtg (blue) and Hoxa1 (red). In the medial neural plate, Hoxa1 and Prtg were coexpressed in the same anteroposterior (AP) domain giving rise to a brown staining. Laterally, cells expressing Prtg (blue cells) were present anterior to the Hoxa1 domain, near the borders of the neural plate. F-H: ISH on mouse embryos at the 7.75 dpc (F), 7.95 dpc (G), and 8 dpc (H) stages, with a Prtg probe. I-K: ISH on mouse embryos at the 7.25 dpc (I), 7.95 dpc (J), and 8 dpc (K) stages, with a Punc probe. L-N: ISH on chick embryos at stages 4 (L), 7 (M), and 8 (N), with a chick Prtg probe. O-Q: ISH on zebrafish embryos at the 100% epiboly (O), 1 s (P), and 4 s (Q) stages, with a zebrafish prtga probe. All embryos are oriented with anterior to the left. In B-E, G, H, J, and K, mouse embryos have been cut posterior to the node. n, node; nf, neural fold; ps, primitive streak; r, rhombomere; s1, somite 1; r3/r4 indicates the prospective boundary between rhombomeres 3 and 4 of the hindbrain.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Dev. Dyn.