|
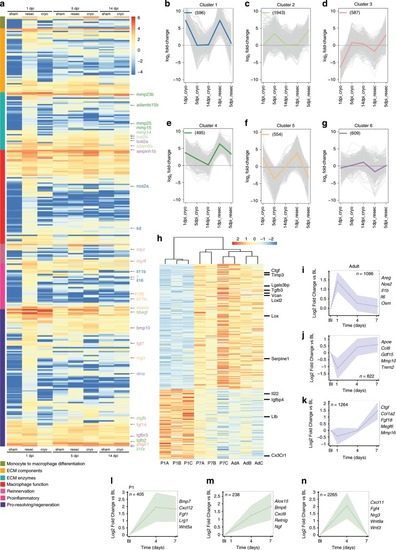
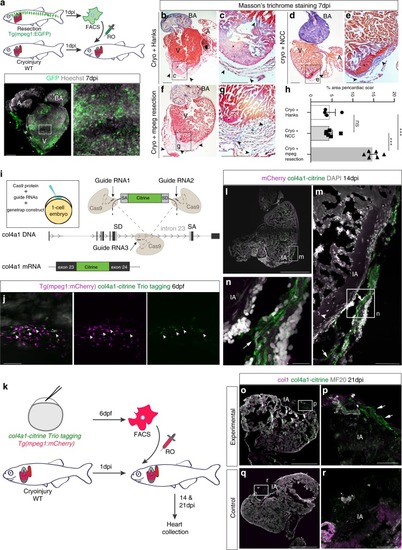
Zebrafish macrophages directly contribute to transient scar formation by collagen synthesis.a Adoptive transfer of GFP+ macrophages derived from a resection heart into a cryoinjured heart via retro-orbital (RO) injection. Wholemount heart showing transplanted macrophages in 7 days post injury (dpi) recipient hearts, hoechst-stained nuclei in grey. Scale bars: 200μm, high-magnification 100 μm. b–g Masson’s trichrome staining of Hanks-injected (b; high-magnification in c), neural crest cells (NCC)-transplanted (d; high-magnification in e), and macrophages-transplanted (f; high-magnification in g) hearts showing pericardiac fibrosis (blue, arrowheads) around the cryoinjured area (pink, black asterisk). Scale bars: 200 μm, high-magnification: 50 μm. h Quantification of scar fibrosis (percentage of total section area, 6 sections per heart: hanks-injected 4.611 ± 1.26%, n = 4 vs macrophage-transplanted 14.87 ± 0.81%, n = 6; ***p < 0.0001; NCC-transplanted 4.53 ± 0.643%, n = 6 vs macrophage-transplanted 14.87 ± 0.81%, n = 6; ***p < 0.0001; hanks-injected 4.611 ± 1.26%, n = 4 vs NCC-transplanted 4.53 ± 0.643%, n = 6; p = 0.951, ns, not significant). 2-tailed, unpaired Student’s t-test, data are mean ± SEM. i “Trio” tagging approach: Cas9 protein, guide RNAs and genetrap construct coding for Citrine, are injected in 1-cell stage embryos. Double-strand breaks are generated in the col4a1 intron and the donor construct, resulting in integration of Citrine ORF flanked by splice acceptor (SA) and donor (SD) sites, ultimately ensuing a col4a1-Citrine fusion protein. j Mosaic expression of col4a1-Citrine observed in 6 dpf macrophages (arrowheads) of “trio” tagging-injected embryos. Scale bar: 50 μm. k Adoptive transfer of mCherry+ macrophages, FAC-sorted from “trio” tagging-injected embryos, transplanted into WT cryoinjured heart via RO injection. l–n 14 dpi WT hearts transplanted with col4a1-Citrine mCherry macrophages. Very few transplanted macrophages (m, arrowhead) are still present in the injury region (dashed line and IA in l, IA in m, n). Macrophage-deposited mosaic “green” scar is observed extracellularly, near the injured area (arrows m, n, DAPI-stained nuclei in grey). (o–r) 21 dpi WT transplanted hearts reveal a mosaic scar (arrows), also stained for collagen 1 (fuschia), peripheral to the MF20-negative injury area (dashed line and IA in o, IA in p), not observable in control conditions (q, r). Scale bars l–n: 200 μm, high-magnification: 100 μm). Representative images of n = 3 per group. Source data are provided as a Source Data file.
|