- Title
-
A 190 base pair, TGF-β responsive tooth and fin enhancer is required for stickleback Bmp6 expression
- Authors
- Erickson, P.A., Cleves, P.A., Ellis, N.A., Schwalbach, K.T., Hart, J.C., Miller, C.T.
- Source
- Full text @ Dev. Biol.
|
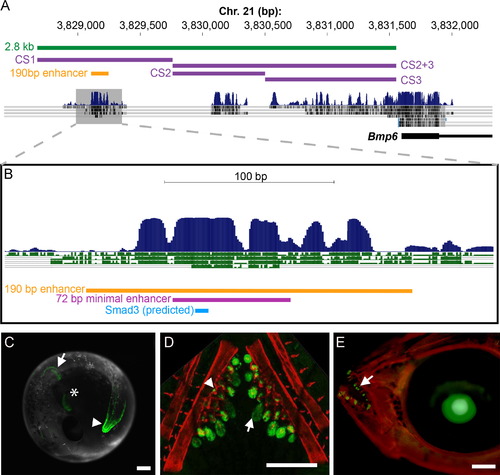
A conserved 190 bp enhancer upstream of Bmp6 drives gene expression in several domains. (A) The 52 region of stickleback Bmp6 from the UCSC genome browser (http://genome.ucsc.edu/). The region of genomic DNA used in the 2.8 kb enhancer construct (see Fig. S3) is shown in green, conserved sequences (CS) 1–3 are shown in purple, and the subcloned 190 bp enhancer is shown in yellow. The first exon and part of the first intron of Bmp6 are shown in thick and thin black lines, respectively (bottom). Conservation peaks and alignments (dark blue and gray) are shown from the 8-Species MultiZ track. (B) Zoom in on the middle of CS1, approximately 2.5 kb upstream of the predicted Bmp6 transcription start site. The 190 bp enhancer, the 72 bp minimal enhancer (see Fig. S6), and a predicted Smad3 binding site (see Fig. 3 and Fig. 4) are shown in yellow, pink, and blue, respectively. The conservation track is shown as dark blue peaks, above green alignments showing conservation to medaka, tetraodon, fugu, and zebrafish, from top to bottom. (C) GFP reporter expression pattern driven by the 190 bp enhancer in a 5 dpf (stage 22, (Swarup, 1958)) stickleback embryo. Strong expression was seen in the distal edge of the developing pectoral fin (arrow), the heart (asterisk), and the distal edge of the median fin (arrowhead). (D) Confocal projection of GFP reporter expression in the ventral pharyngeal tooth plate in a ~10 mm stickleback fry. Expression was observed in the epithelium of developing tooth germs (arrow) and the odontogenic mesenchyme (arrowhead) in the cores of ossified teeth. Bones are fluorescently stained with Alizarin red. (E) GFP reporter expression in the oral teeth (arrow) of a 30 dpf stickleback fry. GFP in the lens is an internal control for the zebrafish hsp70 promoter used in the transgenic construct. Scale bars=200 µm. |
|
Evolutionary functional conservation of the Bmp6 enhancer in teleosts. (A) Sequence alignments of four teleost sequences relative to the 190 bp stickleback enhancer. The perfectly conserved Smad3 dimer binding site is marked in blue, and purple arrows mark the boundaries of the 72 bp minimal enhancer (see Fig. S6). (B-D) The stickleback sequence reporter construct stably integrated into the zebrafish genome drove expression in the distal edge of the median fin at 24 hpf (arrow in B), the distal edge of the pectoral fin at 48 hpf (arrow in C), and tooth epithelium (arrow) and mesenchyme (arrowhead) at 5 dpf (D). (E-G) A 477 bp construct of zebrafish genomic sequence centered around the conserved sequence of the enhancer drove similar, but weaker expression in the median fin of a 33 hpf zebrafish (arrow in E), pectoral fins of a 48 hpf zebrafish (inset of F), and teeth of a 5 dpf zebrafish (G). (H-I) Although not detected in seven of eight stable lines, in one of eight stable lines, the zebrafish sequence drove faint expression in the distal edges of the median fin (arrow in H) and pectoral fins (arrow in I) of 5 dpf stickleback. However, no expression was detected in tooth germs in newly hatched fry in any line (J). See Table S2 for quantification of expression domains of transgenic lines. Bone is fluorescently stained with Alizarin red in (D, G, J). Scale bars=200 µm. |
|
Pharmacological disruption of TGFβ signaling or TALEN-induced mutations of the predicted Smad3 binding site reduce enhancer activity. (A–C) Treatment of stickleback fry for 7 days in SB431542 (an ALK5 inhibitor) severely reduced GFP expression driven by the 190 bp enhancer in a dose-dependent manner. Expression was severely reduced in the epithelia (arrows), but not mesenchyme (asterisks), of pharyngeal teeth at both low (25 µM, B) and high (50 µM, C) doses relative to controls (A). (D-F) SB431542 also eliminated GFP driven by the stickleback enhancer in a zebrafish trans environment. (G) The sequence targeted by TALENs contains a predicted Smad3 homodimer binding site (blue). The TALEN binding sites are indicated in purple text and the purple scissors indicate the approximate site of endonuclease activity. The XbaI site used for molecular screening is underlined in green, and the mutagenized sequence of the Smad3 binding site, indicated by orange letters, is shown below. (H-I) Injection of the TALENs into stable transgenic fish carrying the 190 bp reporter construct resulted in near complete loss of GFP expression in 95% of injected animals (I) relative to controls (H). Residual GFP seen in (I) is likely the result of the mosaicism of TALEN-induced lesions. (J). Mutating the predicted Smad3 binding site resulted in a loss of GFP expression in both epithelium and mesenchyme of pharyngeal teeth in 3/3 stickleback lines observed. Bone is fluorescently counterstained with Alizarin red. Scale bars=200 µm. |
|
Wnt signaling is not required for enhancer function, but Wnt and TGFβ are required for tooth development. Newly hatched stickleback fry were treated with DMSO (control, A), SB431542 (B–C), XAV939 (D–E), or a combination of the two drugs at low (25 µM for SB431542 and 5 µM for XAV939, F) or high (50 µM for SB431542 or 10 µM XAV939, G) doses for 5 days. Main panels show Alizarin red and GFP for the ventral tooth plate; insets show GFP only for mesenchyme of a single tooth from the dorsal tooth plate. (B, C) SB431542 reduced GFP in tooth epithelia (arrows) relative to control (A, and see Fig. 3). However, mesenchymal GFP (arrowhead, inset) was less severely reduced. (D, E) XAV939 alone did not affect GFP expression in epithelia (arrows) or mesenchyme (arrowheads) at either dose. (F, G) No strong additional effect on GFP expression was seen when XAV939 and SB431542 were combined, though mesenchymal GFP appeared slightly lower in the combined dose. (H) A combination of SB431542 and XAV939 significantly reduced ventral pharyngeal tooth number. (I) Treatment with SB431542, but not XAV939, decreased the number of green tooth epithelia relative to total ventral teeth (ratio is expressed as a decimal). XAV939 had no additional effect on green epithelia in combination with SB431542. Tukey HSD P-values of relevant comparisons are shown above with asterisks (*=P<0.05, ** =P<0.0005, n.s.=P>0.05). Scale bars=200 µm. |
|
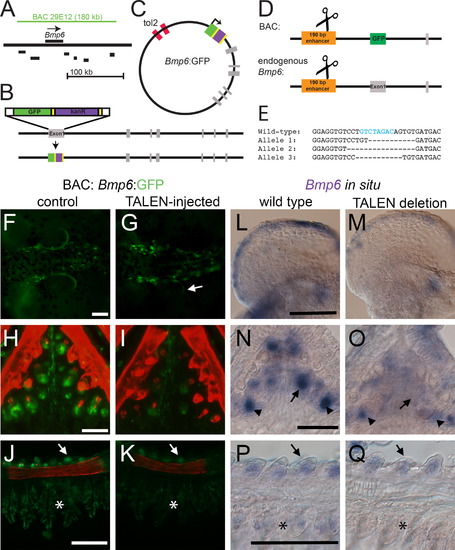
The 5′ 190 bp enhancer is necessary for Bmp6 expression. (A) Schematic of the genomic location of the 180 kb CHORI BAC29E12 with respect to Bmp6 and nearby genes (coding regions shown in black are Ipo4, Pdcd6, Txndc5, Muted, Eef1e1, and Slc35b3 from left to right). (B) Recombineering strategy for introducing GFP into the first exon of Bmp6; gray bars indicate exons. (C) Final circular BAC with inverted Tol2 sites for transposition and GFP reporter (not to scale). (D) Strategy for introducing TALEN lesions into the 190 bp 5′ enhancer. The same TALENs were used to target the enhancer in stable transgenic BAC fish and at the endogenous Bmp6 locus (diagram not to scale). (E) Sequences of stable mutant enhancer alleles. For the endogenous locus targeting, F2 fish trans-heterozygous for two different enhancer mutations were generated. Fish in (M) carried alleles 1 and 2; fish in (O) and (Q) carried alleles 1 and 3. The predicted Smad3 binding site is indicated with blue text in the wild type sequence. (F, G) In the reporter BAC, TALEN injection frequently severely reduced GFP expression from the pectoral fin relative to controls at 5 dpf. A small patch of mosaic, unaffected GFP is indicated with the arrow in (G). (H, I) TALEN injection also eliminated much of the Bmp6 tooth expression (I). (J, K) GFP expression was also reduced in gills (asterisk) and slightly reduced in the gill rakers (arrowhead). (L-M). Mutations in the enhancer caused a reduction in pectoral fin expression relative to wild-type siblings. (N, O) Bmp6 expression was also lost in tooth epithelia (arrows), but was not entirely lost in mesenchyme (arrowheads). (P, Q) Expression was also noticeably reduced in gills (asterisk), though gill raker expression (arrows) appears similar to wild-type sibling controls. Scale bars=100 µm. |
|
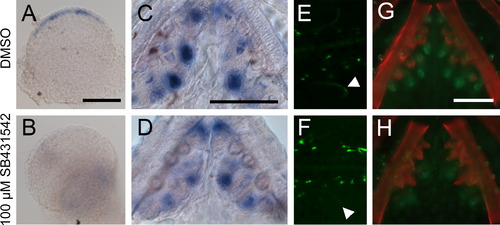
Treatment with TGFβ inhibitor SB431542 reduces Bmp6 expression. (A-D) Sticklebacks were treated with 100 µM SB431542 or vehicle control from 2 to 5 dpf or for 5 days post-hatching, and Bmp6 expression was assayed by in situ hybridization. Drug treatment severely reduced Bmp6 expression in fins (A, B) and also reduced Bmp6 expression in tooth epithelia (C, D). Likewise, GFP driven by the Bmp6 locus in the reporter BAC was also reduced in fins (arrowheads in E, F) and teeth (G, H) after SB431542 treatment. Scale bars=100 µm. |
Reprinted from Developmental Biology, 401(2), Erickson, P.A., Cleves, P.A., Ellis, N.A., Schwalbach, K.T., Hart, J.C., Miller, C.T., A 190 base pair, TGF-β responsive tooth and fin enhancer is required for stickleback Bmp6 expression, 310-23, Copyright (2015) with permission from Elsevier. Full text @ Dev. Biol.